Usutu Virus: An Emerging Flavivirus in Europe
Abstract
:1. Introduction
2. Life Cycle of USUV
| Species | Common Name | Country (year) | References |
|---|---|---|---|
| Dendrocopos major | Great spotted woodpecker | Belgium (2014) | [29] |
| Pyrrhula pyrrhula | Bullfinch | ||
| Columba livia domestica | Domestic pigeon | Greece (2014) | [30] |
| Turdus philomelos | Song thrushes | Spain (2012) | [31] |
| Turdus merula | Eurasian blackbird | Italy (2010–2011) | [18,19,25,26,27,28] |
| Germany (2011) | |||
| Hungary (2003–2006) | |||
| Austria (2001–2005) | |||
| Alcedo atthis | Common kingfisher | Germany (2011) | [26] |
| Serinus canaria domestica | Canary | ||
| Alectoris rufa | Partridge | Italy (2010–2011) | [18,19] |
| Asio otus | Long-eared owl | ||
| Caprimulgus europaeus | Nightjar | ||
| Garrulus glandarius | Eurasian jay | ||
| Larus michahellis | Yellow-legged gull | ||
| Pica pica | Eurasian magpie | ||
| Streptopelica decaocto | Eurasian collared dove | ||
| Ardea cinerea | Grey heron | Germany (2011) | [18,19,26] |
| Merops apiaster | Eurasian bee-eater | Italy (2010–2011) | |
| Passer domesticus | House sparrow | ||
| Picus viridis | Eurasian green woodpecker | ||
| Sturnus vulgaris | Common starling | ||
| Strix nebulosa | Great grey owl | Germany (2011) | [26,27] |
| Austria (2001–2002) | |||
| Gallus gallus domesticus | Chicken | Italy (2007–2009) | [14,32,33,34] |
| Switzerland (2006–2007) | |||
| England (2006) | |||
| Spheniscus humboldti | Humboldt penguin | Switzerland (2006–2007) | [34] |
| Phoenicopterus ruber | Greater flamingo | ||
| Dacelo novaeguineae | Laughing kookaburra | ||
| Ciconia ciconia | White stork | Austria (2006–2007) | [34] |
| Leptoptilos crumeriiferus | Marabou stork | ||
| Neophron percnopterus | Egyptian vulture | ||
| Bubo bubo | Eurasian eagle owl | ||
| Bubo scandiacus | Snowy owl | ||
| Strix uralensis | Ural owl |
3. USUV Surveillance from Africa to Europe
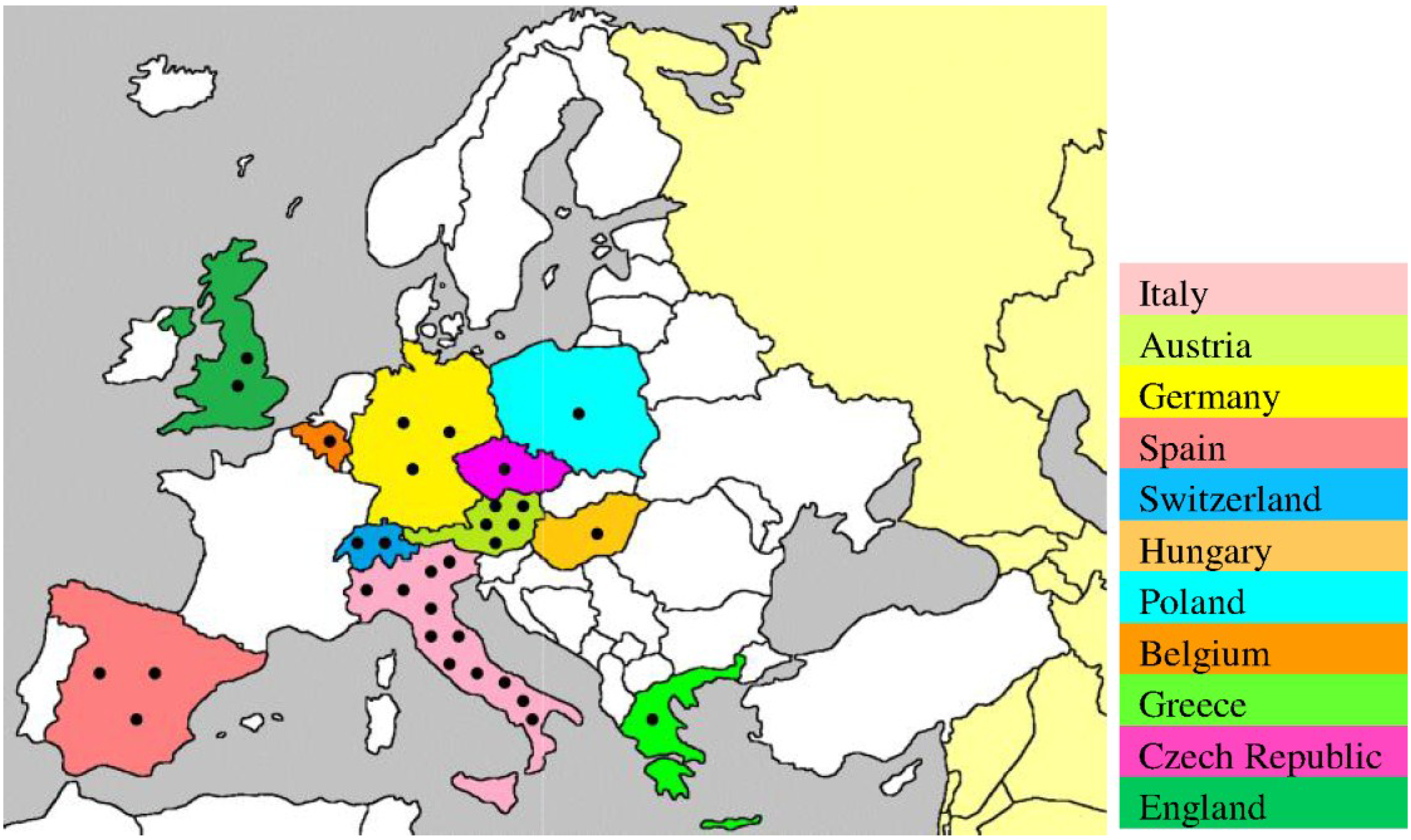
4. USUV Infection in Humans
5. Cellular Tropism and Pathogenesis of USUV
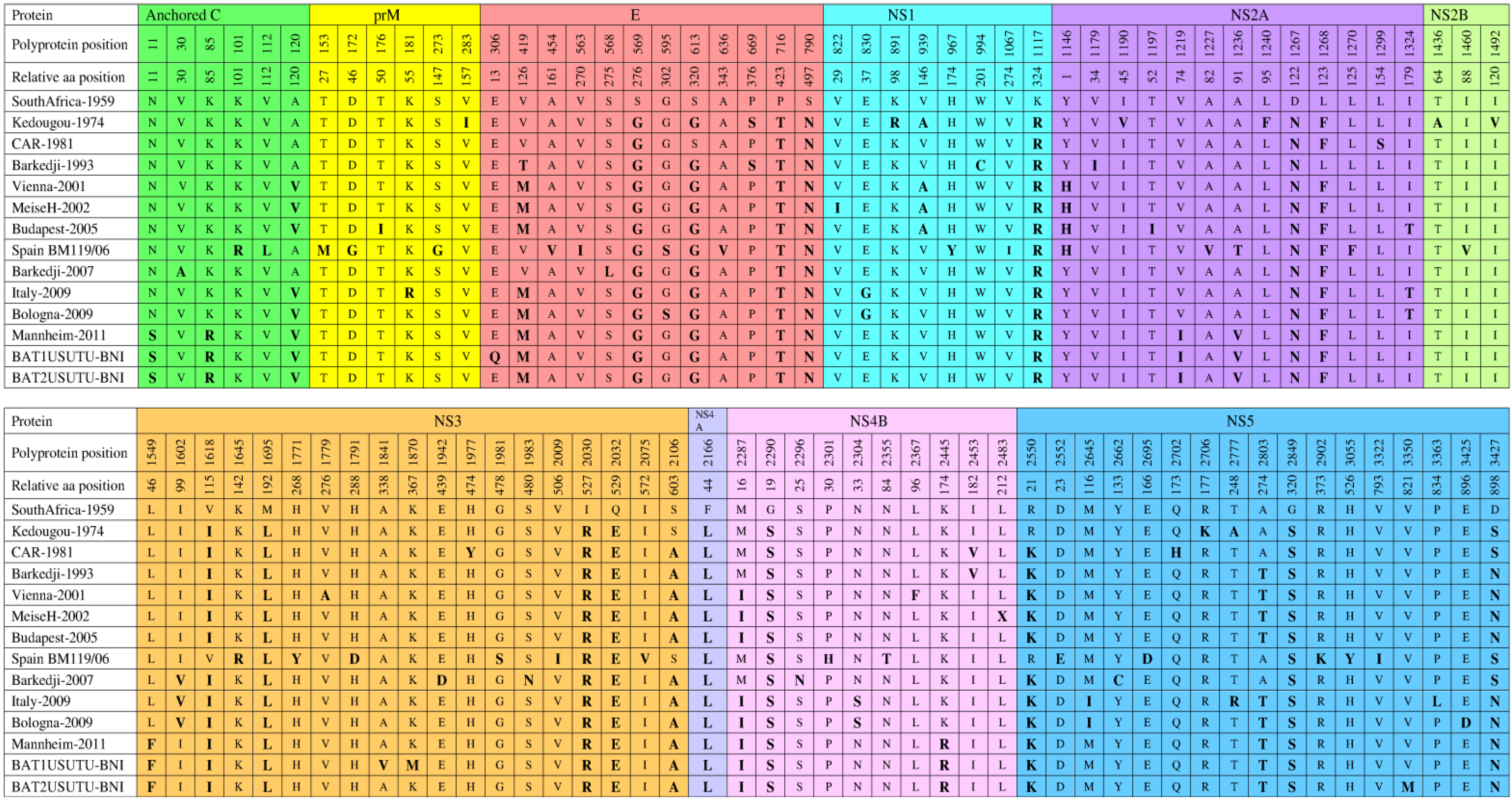
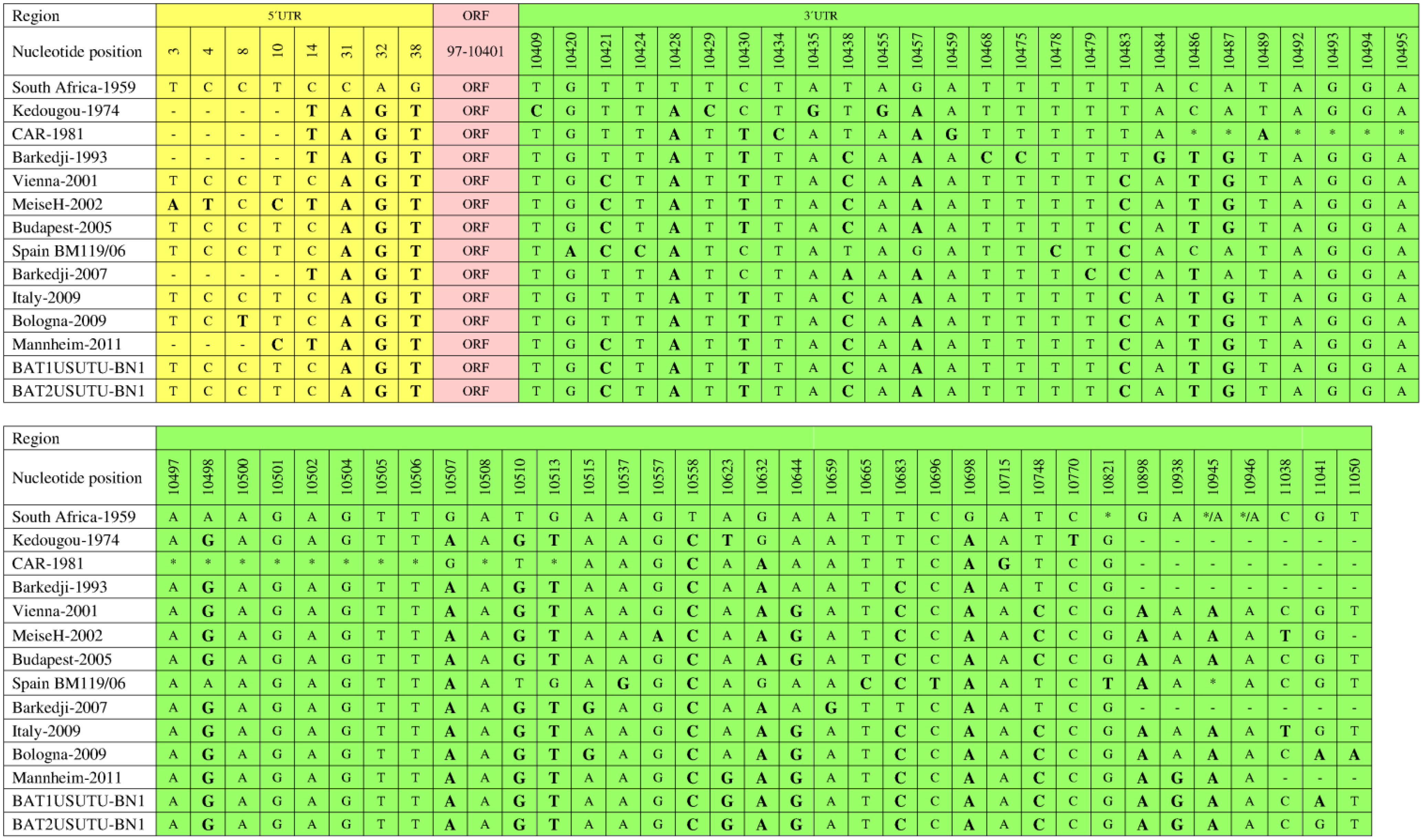
6. Genomic Structure and Phylogenetic Analysis of USUV
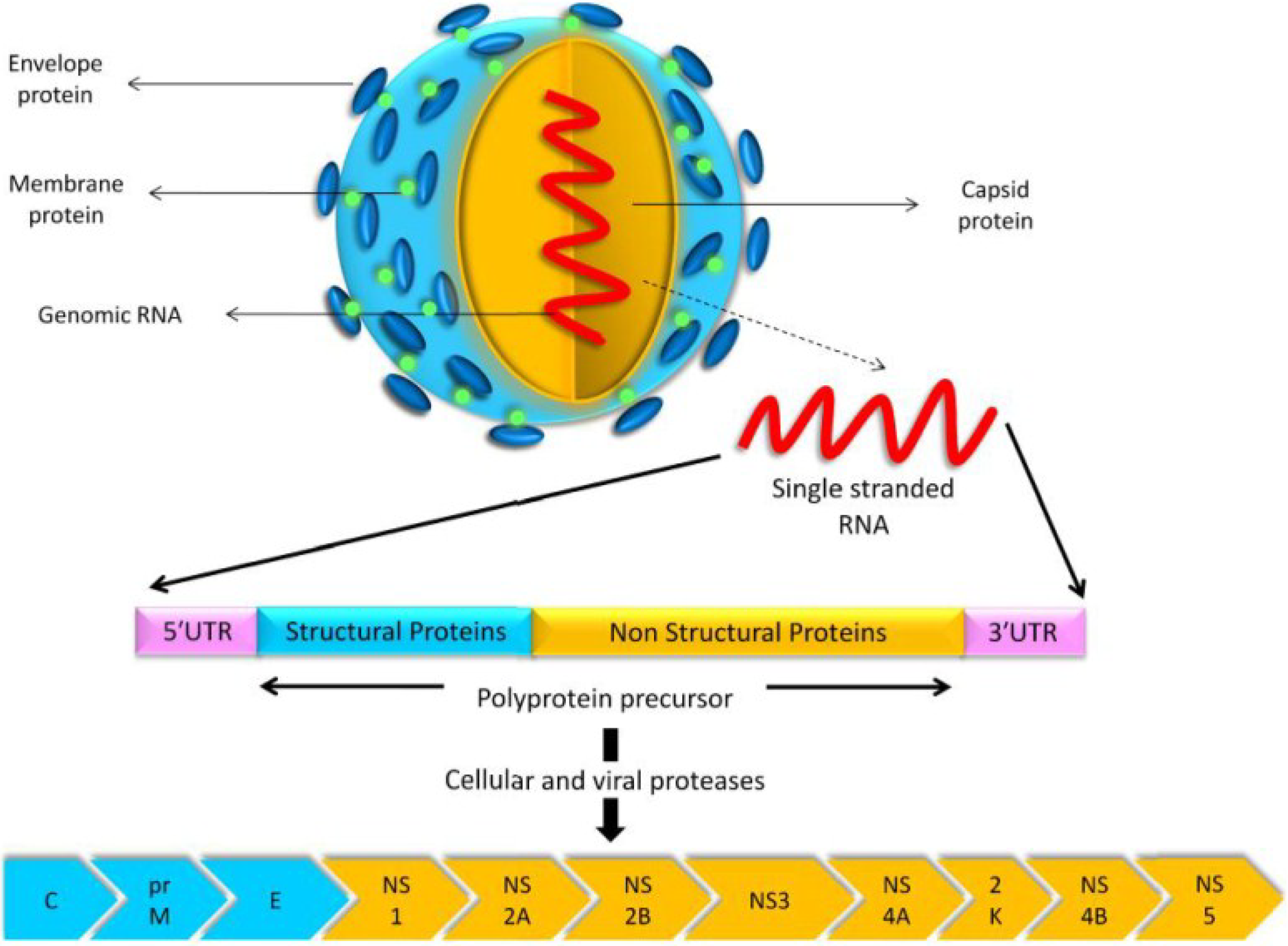
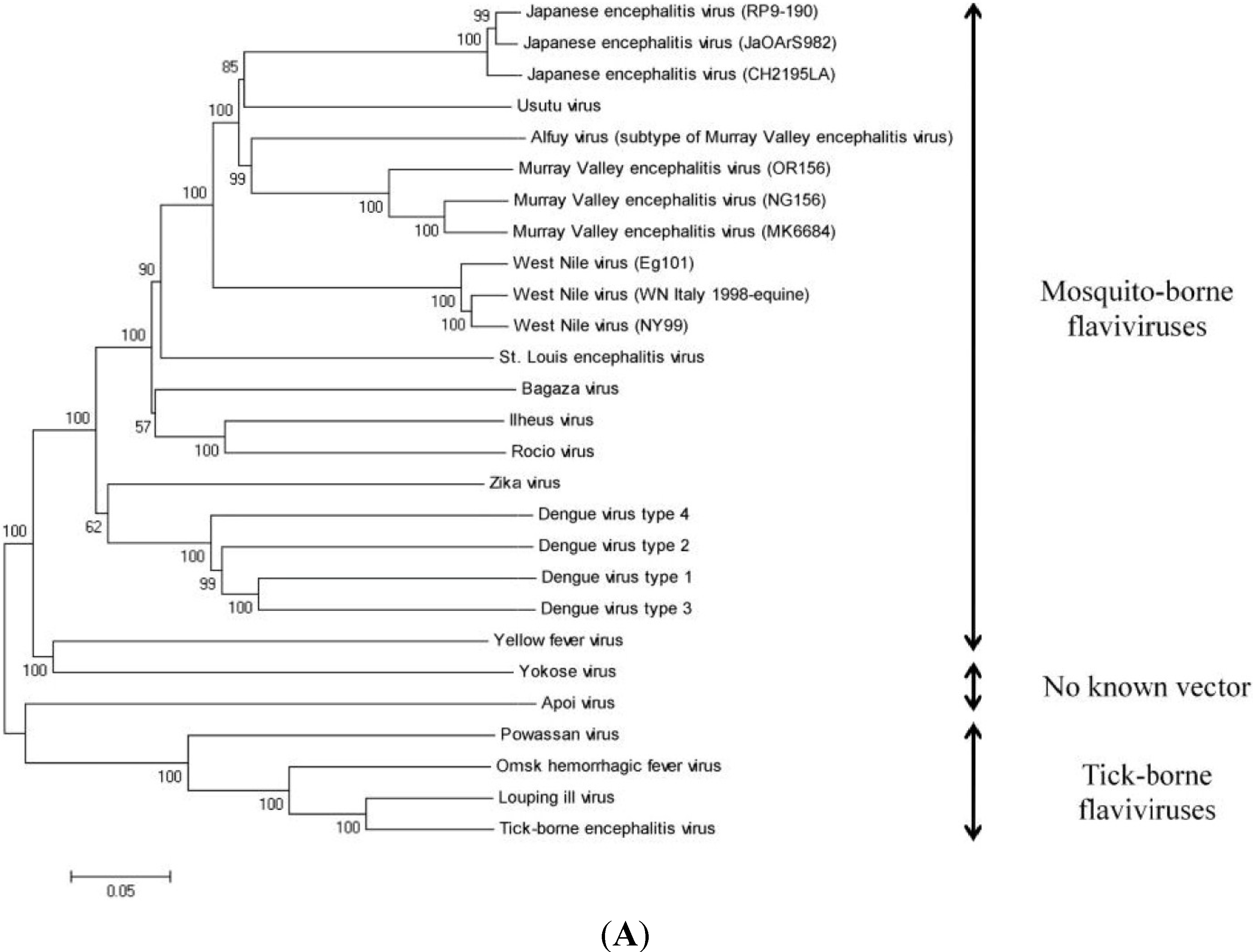
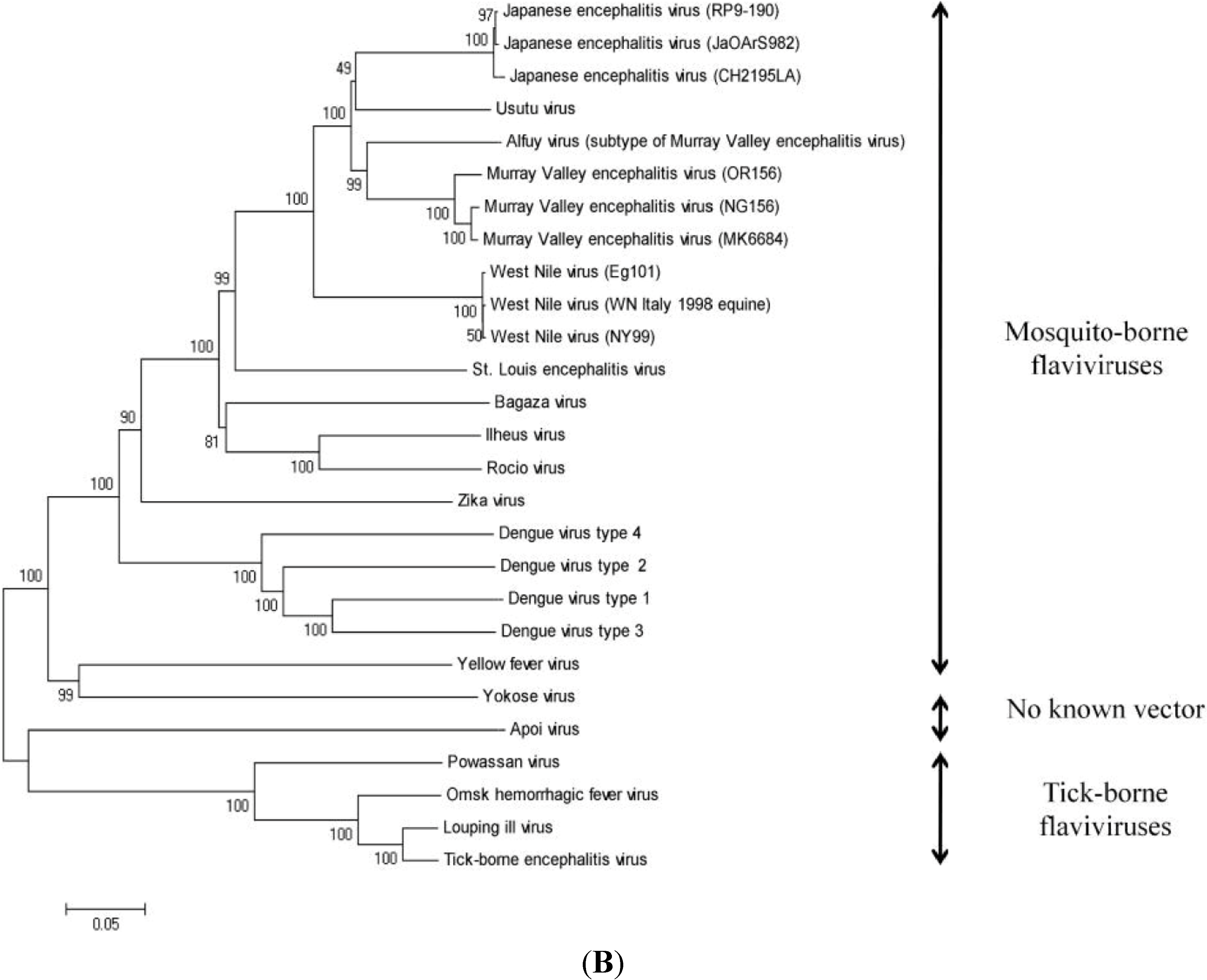
| Virus Name | Nucleotide Accession Number | Protein Accession Number |
|---|---|---|
| Alfuy virus | AY898809 | AAX82481 |
| Apoi virus | NC_003676 | NP_620045 |
| Bagaza virus | HQ644144 | AEI27245 |
| Dengue virus type 1 | AF309641 | AAK62993 |
| Dengue virus type 2 | M29095 | AAA42941 |
| Dengue virus type 3 | AY679147 | AAT79552 |
| Dengue virus type 4 | AF326573 | AAK01233 |
| Ilheus virus | KC481679 | AGJ84083 |
| Japanese encephalitis virus (CH2195LA) | AF221499 | AAF34186 |
| Japanese encephalitis virus (JaOArS982) | M18370 | AAA81554 |
| Japanese encephalitis virus (RP9-190) | KF907505 | AHK05344 |
| Louping ill virus | KF056331 | AGN32859 |
| Murray Valley encephalitis virus (MK6684) | KF751869 | AIA58169 |
| Murray Valley encephalitis virus (NG156) | KF751870 | AIA58170 |
| Murray Valley encephalitis virus (OR156) | KF751871 | AIA58171 |
| Omsk hemorrhagic fever virus | AY193805 | AAP29989 |
| Powassan virus | L06436 | AAA02739 |
| Rocio virus | AY632542 | AAV34158 |
| St. Louis encephalitis virus | NC_007580 | YP_001008348 |
| Tick-borne encephalitis virus | KF151173 | AGP05331 |
| Usutu virus | AY453412 | AAS59401 |
| West Nile virus (Eg101) | AF260968 | AAG02039 |
| West Nile virus (NY99) | DQ211652 | ABA62343 |
| West Nile virus (WN Italy 1998-equine) | AF404757 | AAM81753 |
| Yellow fever virus | DQ235229 | ABB69689 |
| Yokose virus | NC_005039 | NP_872627 |
| Zika virus | AY632535 | AAV34151 |
7. Genetic Diversity among Different USUV Strains
| Strain | Geographical | Genome | Total | Nucleotide | Amino Acid | Nucleotide | Protein |
|---|---|---|---|---|---|---|---|
| Origin | Length (bp) | Amino Acids | Similarity % | Similarity % | Access. No | Access. No | |
| South Africa-1959 | South Africa | 11064 | 3434 | - | - | AY453412 | AAS59401 |
| CAR-1969 | CAR | 10745 | 3434 | 81 | 95 | KC754958 | AGP50649 |
| Kedougou-1974 | Senegal | 10837 | 3434 | 97 | 99 | KC754954 | AGP50645 |
| CAR-1981 | CAR | 10800 | 3434 | 97 | 99 | KC754955 | AGP50646 |
| Barkedji-1993 | Senegal | 10837 | 3434 | 97 | 99 | KC754956 | AGP50647 |
| Vienna-2001 | Austria | 11066 | 3434 | 97 | 99 | AY453411 | AAS59402 |
| MeiseH-2002 | Austria | 11047 | 3434 | 97 | 99 | JQ219843 | AFE85504 |
| Budapest-2005 | Hungary | 11065 | 3434 | 97 | 99 | EF206350 | ABP88817 |
| Spain BM119/06 | Spain | 11064 | 3434 | 97 | 99 | KF573410 | AHA57377 |
| Barkedji-2007 | Senegal | 10825 | 3434 | 97 | 99 | KC754957 | AGP50648 |
| Italy-2009 | Italy | 11065 | 3434 | 97 | 99 | JF266698 | AEK21245 |
| Bologna-2009 | Italy | 11065 | 3434 | 97 | 99 | HM569263 | AEF13245 |
| Mannheim-2011 | Germany | 11003 | 3434 | 97 | 99 | HE599647 | CCD57503 |
| BAT1USUTU-BNI | Germany | 11065 | 3434 | 97 | 99 | KJ859682 | AIN76231 |
| BAT2USUTU-BNI | Germany | 11065 | 3434 | 97 | 99 | KJ859683 | AIN76232 |
8. Diagnosis
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Karabatsos, N. Usutu virus. In International Catalogue of Arboviruses Including Certain other Viruses of Vertebrates, 3rd ed.; The American Society of Tropical Medicine and Hygiene: San Antonio, TX, USA, 1985; pp. 1059–1060. [Google Scholar]
- Kuno, G.; Chang, G.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [PubMed]
- Poidinger, M.; Hall, R.A.; Mackenzie, J.S. Molecular characterization of the Japanese encephalitis serocomplex of the Flavivirus genus. Virology 1996, 218, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Gould, E.A. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 2003, 59, 1–19. [Google Scholar] [PubMed]
- Woodall, J.P. The viruses isolated from arthropods at the East African virus research institute in the 26 years ending December 1963. Proc. E. Afr. Acad. 1964, 2, 141–146. [Google Scholar]
- Adam, F.; Digoutte, J.-P. Virus d’ Afrique (base de donnes). Centre Collaborateur OMS de Référence et de Recherche Pour les Arbovirus et les Virus de Fièvreshémorrhagiques (CRORA); Institut Pasteur de Dakar: Dakar, Senegal. Available online: http://www.pasteur.fr/recherche/banques/CRORA/ (accessed on 12 July 2014).
- Weissenböck, H.; Kolodziejek, J.; Url, A.; Lussy, H.; Rebel-Bauder, B.; Nowotny, N. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, central Europe. Emerg. Infect. Dis. 2002, 8, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Mani, P.; Rossi, G.; Perrucci, S.; Bertini, S. Mortality of Turdus merula in Tuscany. Sel. Vet. 1998, 8, 749–753. (In Italian) [Google Scholar]
- Weissenböck, H.; Bakonyi, T.; Rossi, G.; Mani, P.; Nowotny, N. Usutu virus, Italy, 1996. Emerg. Infect. Dis. 2013, 19, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; Ruiz, S.; Herrero, L.; Moreno, J.; Molero, F.; Magallanes, A.; Sánchez-Seco, M.P.; Figuerola, J.; Tenorio, A. West Nile and Usutu viruses in mosquitoes in Spain, 2008–2009. Am. J. Trop. Med. Hyg. 2011, 85, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, H.W.; Bakonyi, T.; Weissenböck, H.; Hatt, J.M.; Eulenberger, U.; Robert, N.; Hoop, R.; Nowotny, N. Emergence and establishment of Usutu virus infection in wild and captive avian species in and around Zurich, Switzerland–genomic and pathologic comparison to other central European outbreaks. Vet. Microbiol. 2011, 148, 207–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecorari, M.; Longo, G.; Gennari, W.; Grottola, A.; Sabbatini, A.; Tagliazucchi, S.; Savini, G.; Monaco, F.; Simone, M.; Lelli, R.; et al. First human case of Usutu virus neuroinvasive infection, Italy, August-September 2009. Euro. Surveill. 2009, 14, e19446. [Google Scholar]
- Cavrini, F.; Gaibani, P.; Longo, G.; Pierro, A.M.; Rossini, G.; Bonilauri, P.; Gerunda, G.E.; di Benedetto, F.; Pasetto, A.; Girardis, M.; et al. Usutu virus infection in a patient who underwent orthotropic liver transplantation, Italy, August-September 2009. Euro. Surveill. 2009, 14, e19448. [Google Scholar]
- Savini, G.; Monaco, F.; Terregino, C.; di Gennaro, A.; Bano, L.; Pinoni, C.; de Nardi, R.; Bonilauri, P.; Pecorari, M.; di Gialleonardo, L.; et al. Usutu virus in Italy: An emergence or a silent infection? Vet. Microbiol. 2011, 151, 264–274. [Google Scholar] [CrossRef]
- Cadar, D.; Becker, N.; Campos Rde, M.; Börstler, J.; Jöst, H.; Schmidt-Chanasit, J. Usutu virus in bats, Germany, 2013. Emerg. Infect. Dis. 2014, 20, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, M.; Bonilauri, P.; Bellini, R.; Albieri, A.; Defilippo, F.; Maioli, G.; Galletti, G.; Gelati, A.; Barbieri, I.; Tamba, M.; et al. Evidence of simultaneous circulation of West Nile and Usutu viruses in mosquitoes sampled in Emilia-Romagna region (Italy) in 2009. PLoS One 2010, 5, e14324. [Google Scholar] [CrossRef]
- Busquets, N.; Alba, A.; Allepuz, A.; Aranda, C.; Ignacio Nuñez, J. Usutu virus sequences in Culex pipiens (Diptera: Culicidae), Spain. Emerg. Infect. Dis. 2008, 14, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, M.; Gaibani, P.; Bellini, R.; Defilippo, F.; Pierro, A.; Albieri, A.; Mailoli, G.; Luppi, A.; Rossini, G.; Balzani, A.; et al. Mosquito, bird, and human surveillance of West Nile and Usutu viruses in Emilia-Romagna region (Italy) in 2010. PLoS One 2012, 7, e38058. [Google Scholar] [CrossRef]
- Calzolari, M.; Bonilauri, P.; Bellini, R.; Albieri, A.; Defilippo, F.; Tamba, M.; Tassinari, M.; Gelati, A.; Cordioli, P.; Angelini, P.; et al. Usutu virus persistence and West Nile virus inactivity in the Emilia-Romagna region (Italy) in 2011. PLoS One 2013, 8, e63978. [Google Scholar] [CrossRef]
- Tamba, M.; Bonilauri, P.; Bellini, R.; Calzolari, M.; Albieri, A.; Sambri, V.; Dottori, M.; Angelini, P. Detection of Usutu virus within a West Nile virus surveillance program in northern Italy. Vector Borne Zoonotic Dis. 2011, 11, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Jöst, H.; Bialonski, A.; Maus, D.; Sambri, V.; Eiden, M.; Groschup, M.H.; Günther, S.; Becker, N.; Schmidt-Chanasit, J. Isolation of Usutu virus in Germany. Am. J. Trop. Med. Hyg. 2011, 85, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Cornet, M.; Robin, Y.; Chateau, R.; Heme, G.; Adam, C.; Valade, M.; Le Gonidec, G.; Jan, C.; Renaudet, J.; Dieng, P.Y.; et al. Isolementd’arbovirus au Sénégal Oriental a partir de moustiques (1972–1977) et notes surl’épidémiologie des virus transmis par les Aedes en particulier du virus amaril. Cahiers ORSTOM. Sér. Entomologieméd. Parasitol. 1979, 17, 149–163. [Google Scholar]
- Hubálek, Z. Pathogenic microorganisms associated with free-living birds (a review). Acta Sci. Nat. Brno. 1994, 28, 1–74. [Google Scholar]
- Nikolay, B.; Diallo, M.; Faye, O.; Boye, C.S.; Sall, A.A. Vector competence of Culex neavei (Diptera: Culicidae) for Usutu virus. Am. J. Trop. Med. Hyg. 2012, 86, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Erdélyi, K.; Ursu, K.; Ferenczi, E.; Csörgo, T.; Lussy, H.; Chvala, S.; Bukovsky, C.; Meister, T.; Weissenböck, H.; et al. Emergence of Usutu virus in Hungary. J. Clin. Microbiol. 2007, 45, 3870–3874. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Jöst, H.; Ziegler, U.; Eiden, M.; Höper, D.; Emmerich, P.; Fichet-Calvet, E.; Ehichioya, D.U.; Czajka, C.; Gabriel, M.; et al. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS One 2012, 7, e32604. [Google Scholar] [CrossRef] [PubMed]
- Weissenböck, H.; Kolodziejek, J.; Fragner, K.; Kuhn, R.; Pfeffer, M.; Nowotny, N. Usutu virus activity in Austria, 2001–2002. Microbes Infect. 2003, 5, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Chvala, S.; Bakonyi, T.; Bukovsky, C.; Meister, T.; Brugger, K.; Rubel, F.; Nowotny, N.; Weissenböck, H. Monitoring of Usutu virus activity and spread by using dead bird surveillance in Austria, 2003–2005. Vet. Microbiol. 2007, 122, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.M.; Marlier, D.; Tenner-Racz, K.; Eiden, M.; Cassart, D.; Gandar, F.; Beer, M.; Schmidt-Chanasit, J.; Desmecht, D. Detection of Usutu virus in a bullfinch (Pyrrhula pyrrhula) and a great spotted woodpecker (Dendrocopos major) in north-west Europe. Vet. J. 2014, 199, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Chaintoutis, S.C.; Dovas, C.I.; Papanastassopoulou, M.; Gewehr, S.; Danis, K.; Beck, C.; Lecollinet, S.; Antalis, V.; Kalaitzopoulous, S.; Panagiotopoulos, T.; et al. Evaluation of a West Nile virus surveillance and early warning system in Greece, based on domestic pigeons. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 131–141. [Google Scholar] [CrossRef]
- Höfle, U.; Gamino, V.; de Mera, I.G.; Mangold, A.J.; Ortίz, J.A.; de la Fuente, J. Usutu virus in migratory song thrushes, Spain. Emerg. Infect. Dis. 2013, 19, 1173–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, A.; Dawson, A.; Gould, E.A. Detection of seroconversion to West Nile virus, Usutu virus and Sindbis virus in UK sentinel chickens. Virol. J. 2006, 3, e71. [Google Scholar] [CrossRef] [Green Version]
- Steinmetz, H.W.; Bakonyi, T.; Chvala, S.; Weissenböck, H.; Eulenberger, U.; Hatt, J.M.; Hoop, R.; Nowotny, N. Emergence of Usutu virus in Switzerland. In Proceedings of the 43rd International Symposium on Diseases of Zoo and Wild Animals, Edinburgh, UK, 16–20 May 2007; pp. 129–131.
- Buchebner, N.; Zenker, W.; Wenker, C.; Steinmetz, H.W.; Sós, E.; Lussy, H.; Nowotny, N. Low Usutu virus seroprevalence in four zoological gardens in central Europe. BMC Vet. Res. 2013, 9, e153. [Google Scholar] [CrossRef]
- Nikolay, B.; Diallo, M.; Boye, C.S.; Sall, A.A. Usutu virus in Africa. Vector Borne Zoonotic Dis. 2011, 11, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, V.; Reynaud, P.; Lefrancois, T.; Durand, B.; Baillon, F.; Balanca, G.; Gaidet, N.; Mondet, B.; Lancelot, R. Predicting West Nile virus seroprevalence in wild birds in Senegal. Vector Borne Zoonotic Dis. 2009, 9, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Ben Hassine, T.; de Massis, F.; Calistri, P.; Savini, G.; BelHaj Mohamed, B.; Ranen, A.; di Gennaro, A.; Sghaier, S.; Hammami, S. First detection of co-circulation of West Nile and Usutu viruses in equids in the south-west of Tunisia. Transbound. Emerg. Dis. 2014, 61, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Manarolla, G.; Bakonyi, T.; Gallazzi, D.; Crosta, L.; Weissenböck, H.; Dorrestein, G.M.; Nowotny, N. Usutu virus in wild birds in northern Italy. Vet. Microbiol. 2010, 141, 159–163. [Google Scholar]
- Hubálek, Z.; Halouzka, J.; Juricová, Z.; Sikutová, S.; Rudolf, I.; Honza, M.; Janková, J.; Chytil, J.; Marec, F.; Sitko, J. Serologic survey of birds for West Nile flavivirus in southern Moravia (Czech Republic). Vector Borne Zoonotic Dis. 2008, 8, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Figuerola, J.; Soriguer, R.; Rojo, G.; Gómez Tejedor, C.; Jimenez-Clavero, M.A. Seroconversion in wild birds and local circulation of West Nile virus, Spain. Emerg. Infect. Dis. 2007, 13, 1915–1917. [Google Scholar] [CrossRef] [PubMed]
- Hubálek, Z.; Wegner, E.; Halouzka, J.; Tryjanowski, P.; Jerzak, L.; Sikutová, S.; Rudolf, I.; Kruszewicz, A.G.; Jaworski, Z.; Wlodarczky, R. Serologic survey of potential vertebrate hosts for West Nile virus in Poland. Viral Immunol. 2008, 21, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Linke, S.; Niedrig, M.; Kaiser, A.; Ellerbrok, H.; Müller, K.; Müller, T.; Conraths, F.J.; Mühle, R.U.; Schmidt, D.; Köppen, U.; et al. Serologic evidence of West Nile virus infections in wild birds captured in Germany. Am. J. Trop. Med. Hyg. 2007, 77, 358–364. [Google Scholar] [PubMed]
- Lelli, R.; Savini, G.; Teodori, L.; Filipponi, G.; Di Gennaro, A.; Leone, A.; Gialleonardo, L.; Venturi, L.; Caporate, V. Serological evidence of Usutu virus occurrence in north-eastern Italy. Zoonoses Public Health 2008, 55, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Hubálek, Z.; Rudolf, I.; Čapek, M.; Bakonyi, T.; Betášová, L.; Nowotny, N. Usutu virus in blackbirds (Turdus merula), Czech Republic, 2011–2012. Transbound. Emerg. Dis. 2014, 61, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; Jiménez-Clavero, M.A.; Franco, L.; Donoso-Mantke, O.; Sambri, V.; Niedrig, M.; Zeller, H.; Tenorio, A. Usutu virus–potential risk of human disease in Europe. Euro. Surveill. 2011, 16, e19935. [Google Scholar]
- Nikolay, B.; Dupressoir, A.; Firth, C.; Faye, O.; Boye, C.S.; Diallo, M.; Sall, A.A. Comparative full length genome sequence analysis of Usutu virus isolates from Africa. Virol. J. 2013, 10, e217. [Google Scholar] [CrossRef]
- Gaibani, P.; Cavrini, F.; Gould, E.A.; Rossini, G.; Pierro, A.; Landini, M.P.; Sambri, V. Comparative genomic and phylogenetic analysis of the first Usutu virus isolate from a human patient presenting with neurological symptoms. PLoS One 2013, 8, e64761. [Google Scholar] [CrossRef] [PubMed]
- Allering, L.; Jöst, H.; Emmerich, P.; Günther, S.; Lattwein, E.; Schmidt, M.; Seifried, E.; Sambri, V.; Hourfar, K.; Schmidt-Chanasit, J. Detection of Usutu virus infection in a healthy blood donor from south-west Germany, 2012. Euro. Surveill. 2012, 17, e20341. [Google Scholar]
- Gaibani, P.; Pierro, A.; Alicino, R.; Rossini, G.; Cavrini, F.; Landini, M.P.; Sambri, V. Detection of Usutu virus-specific IgG in blood donors from northern Italy. Vector Borne Zoonotic Dis. 2012, 12, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Vilibic-Cavlek, T.; Kaic, B.; Barbic, L.; Pem-Novosel, I.; Slavic-Vrzic, V.; Lesniker, V.; Kurecic-Filipovic, S.; Babic-Erceg, A.; Listes, E.; Stevanovic, V.; et al. First evidence of simultaneous occurrence of West Nile virus and Usutu virus neuroinvasive disease in humans in Croatia during the 2013 outbreak. Infection 2014, 42, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Lussy, H.; Weissenböck, H.; Hornyák, A.; Nowotny, N. In vitro host-cell susceptibility to Usutu virus. Emerg. Infect. Dis. 2005, 11, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Brassil, M.M.; Blahnik, G.; McMillan, A.; Ramos, H.J.; Proll, S.C.; Belisle, S.E.; Katze, M.G.; Gale, M., Jr. A systems biology approach reveals that tissue tropism to West Nile virus is regulated by antiviral genes and innate immune cellular processes. PLoS Pathog. 2013, 9, e1003168. [Google Scholar] [CrossRef] [PubMed]
- Weissenböck, H.; Bakonyi, T.; Chvala, S.; Nowotny, N. Experimental Usutu virus infection of suckling mice causes neuronal and glial cell apoptosis and demyelination. Acta Neuropathol. 2004, 108, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Chvala, S.; Kolodziejek, J.; Nowotny, N.; Weissenböck, H. Pathology and viral distribution in fetal Usutu virus infection of birds from the 2001 and 2002 outbreaks in Austria. J. Comp. Pathol. 2004, 131, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.N.; Kent, K.A.; Bennett, C.J.; Bernard, K.A. Tissue tropism and neuroinvasion of West Nile virus do not differ for two mouse strains with different survival rates. Virology 2007, 368, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; Levine, B. Viral evasion of autophagy. Autophagy 2008, 4, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Khakpoor, A.; Panyasrivanit, M.; Wikan, N.; Smith, D.R. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J. Gen. Virol. 2009, 90, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Liang, J.J.; Liao, C.L.; Lin, Y.L. Autophagy is involved in the early step of Japanese encephalitis virus infection. Microbes Infect. 2012, 14, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, A.B.; Escribano-Romero, E.; Merino-Ramos, T.; Saiz, J.S.; Martín-Acebes, M.A. Infection with Usutu virus induces an autophagic response in mammalian cells. PLoS Negl. Trop. Dis. 2013, 7, e2509. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Hsu, Y.W.; Liao, C.L.; Lin, Y.L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2006, 80, 11868–11880. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Gould, E.A.; Kolodziejek, J.; Weissenböck, H.; Nowotny, N. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: Comparison with the South African strain SAAR-1776 and other flaviviruses. Virology 2004, 328, 301–310. [Google Scholar] [PubMed]
- Fulop, L.D.; Barrett, A.D.; Titball, R.W. Nucleotide sequence of the NS5 gene of Banzi virus: Comparison with other flaviviruses. J. Gen. Virol. 1995, 76, 2317–2321. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.F.; Hussain, M.H.; Reid, H.W.; Gould, E.A. Classification of a new member of the TBE flavivirus subgroup by its immunological, pathogenetic and molecular characteristics: Identification of subgroup-specific pentapeptides. Virus Res. 1993, 30, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Peletto, S.; lo Presti, A.; Modesto, P.; Cella, E.; Acutis, P.L.; Farchi, F.; Coitti, M.; Zehender, G.; Ciccozzi, M. Genetic diversity of Usutu virus. New Microbiol. 2012, 35, 167–174. [Google Scholar] [PubMed]
- Solomon, T.; Dung, M.; Vaughn, D.W.; Kneen, R.; Thao, L.T.; Raengsakulrach, B.; Loan, H.T.; Day, N.P.; Farrar, J.; Myint, K.S. Neurological manifestations of dengue infection. Lancet 2000, 25, 1053–1059. [Google Scholar] [CrossRef]
- McMinn, P.C. The molecular basis of virulence of the encephalitogenic flaviviruses. J. Gen. Virol. 1997, 78, 2711–2722. [Google Scholar] [PubMed]
- Chu, J.J.; Rajamanonmani, R.; Bhuvanakantham, R.; Lescar, j.; Ng, M.L. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J. Gen. Virol. 2005, 86, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Halevy, M.; Nestorowicz, A.; Rice, C.M.; Lusting, S. West Nile virus envelope proteins: Nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J. Gen. Virol. 1998, 79, 2375–2380. [Google Scholar] [PubMed]
- Liu, W.J.; Wang, X.J.; Mokhonov, V.V.; Shi, P.Y.; Randall, R.; Khromykh, A.A. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 2005, 79, 1934–1942. [Google Scholar] [CrossRef] [PubMed]
- Beasley, D.W.; Barrett, A.D. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 2002, 76, 13097–13100. [Google Scholar] [CrossRef] [PubMed]
- Proutski, V.; Gould, E.A.; Holmes, E.C. Secondary structure of the 3ʹ untranslated region of flaviviruses: Similarities and differences. Nucleic Acids Res. 1997, 25, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.Y.; Li, W.; Brinton, M.A. Cell proteins bind specifically to West Nile virus minus-strand 3ʹ stem-loop RNA. J. Virol. 1996, 70, 6278–6287. [Google Scholar] [PubMed]
- Institut Pasteur de Dakar. Rapport Annuel, 1970.
- Cavrini, F.; Della Pepa, M.E.; Gaibani, P.; Pierro, A.M.; Rossini, G.; Landini, M.P.; Sambri, V. A rapid and specific real-time RT-PCR assay to identify Usutu virus in human plasma, serum, and cerebrospinal fluid. J. Clin. Virol. 2011, 50, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, B.; Weidmann, M.; Dupressoir, A.; Faye, O.; Boye, C.S.; Diallo, M.; Sall, A.A. Development of a Usutu virus specific real-time reverse transcription PCR assay based on sequenced strains from Africa and Europe. J. Virol. Methods 2014, 197, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.A.; Noga, A.; Kosoy, O.; Johnson, A.J.; Petersen, L.R.; Lanciotti, R.S. Evaluation of a diagnostic algorithm using immunoglobulin M enzyme-linked immunosorbent assay to differentiate human West Nile virus and St. Louis encephalitis virus infections during the 2002 West Nile virus epidemic in the United States. Clin. Diagn. Lab. Immunol. 2004, 11, 1130–1133. [Google Scholar] [PubMed]
- Solomon, T. Flavivirus encephalitis. N. Engl. J. Med. 2004, 351, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Meister, T.; Lussy, H.; Bakonyi, T.; Sikutová, S.; Rudolf, I.; Vogl, W.; Winkler, H.; Frey, H.; Hubálek, Z.; Nowotny, N.; et al. Serological evidence of continuing high Usutu virus (Flaviviridae) activity and establishment of herd immunity in wild birds in Austria. Vet. Microbiol. 2008, 127, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Brazil, M. Birds of East Asia: Eastern China, Taiwan, Korea, Japan, Eastern Russia; Christopher Helm: London, UK, 2009; p. 528. [Google Scholar]
- Wang, G.; Li, C.; Guo, X.; Xing, D.; Dong, Y.; Wang, Z.; Zhang, Y.; Liu, M.; Zheng, Z.; Zhang, H.; et al. Identifying the main mosquito species in China based on DNA barcoding. PLoS One 2012, 7, e47051. [Google Scholar] [CrossRef] [PubMed]
- Knight, K.L.; Stone, A. A Catalog of the Mosquitoes of the World (Diptera: Culicidae), 2nd ed.; Thomas Say Foundation: Maryland, USA, 1977; Volume 6, pp. 1–610. [Google Scholar]
- Kim, H.C.; Friendly, O.S.; Pike, J.G.; Schuster, A.L.; O’Guinn, M.L.; Klein, T.A. Seasonal prevalence of mosquito collected from light traps in the Republic of Korea, 2001. Korean J. Entomol. 2003, 33, 189–201. [Google Scholar] [CrossRef]
- Lobigs, M.; Pavy, M.; Hall, R.A.; Lobigs, P.; Cooper, P.; Komiya, T.; Toriniwa, H.; Petrovsky, N. An inactivated Vero cell-grown Japanese encephalitis vaccine formulated with Advax, a novel inulin-based adjuvant, induces protecting neutralizing antibody against homologous and heterologous flaviviruses. J. Gen. Virol. 2010, 91, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Petrovsky, N.; Larena, M.; Siddharthan, V.; Prow, N.A.; Hall, R.A.; Lobigs, M.; Morrey, J. An inactivated cell culture Japanese encephalitis vaccine (JE-ADVAX) formulated with delta inulin adjuvant provides robust heterologous protection against West Nile encephalitis via cross-protective memory B cells and neutralizing antibody. J. Virol. 2013, 87, 10324–10333. [Google Scholar] [CrossRef] [PubMed]
- Merino-Ramos, T.; Blázquez, A.B.; Escribano-Romero, E.; Cañas-Arranz, R.; Sobrino, F.; Saiz, J.C.; Martín-Acebes, M.A. Protection of a single dose West Nile virus recombinant subviral particle vaccine against linage 1 or 2 strains and analysis of the cross-reactivity with Usutu virus. PLoS One 2014, 9, e108056. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, U.; Ye, J.; Ruan, X.; Wan, S.; Zhu, B.; Cao, S. Usutu Virus: An Emerging Flavivirus in Europe. Viruses 2015, 7, 219-238. https://doi.org/10.3390/v7010219
Ashraf U, Ye J, Ruan X, Wan S, Zhu B, Cao S. Usutu Virus: An Emerging Flavivirus in Europe. Viruses. 2015; 7(1):219-238. https://doi.org/10.3390/v7010219
Chicago/Turabian StyleAshraf, Usama, Jing Ye, Xindi Ruan, Shengfeng Wan, Bibo Zhu, and Shengbo Cao. 2015. "Usutu Virus: An Emerging Flavivirus in Europe" Viruses 7, no. 1: 219-238. https://doi.org/10.3390/v7010219





