Role of microRNAs in Arbovirus/Vector Interactions
Abstract
:1. Introduction
2. Biogenesis of miRNAs
2.1. Canonical Pathway
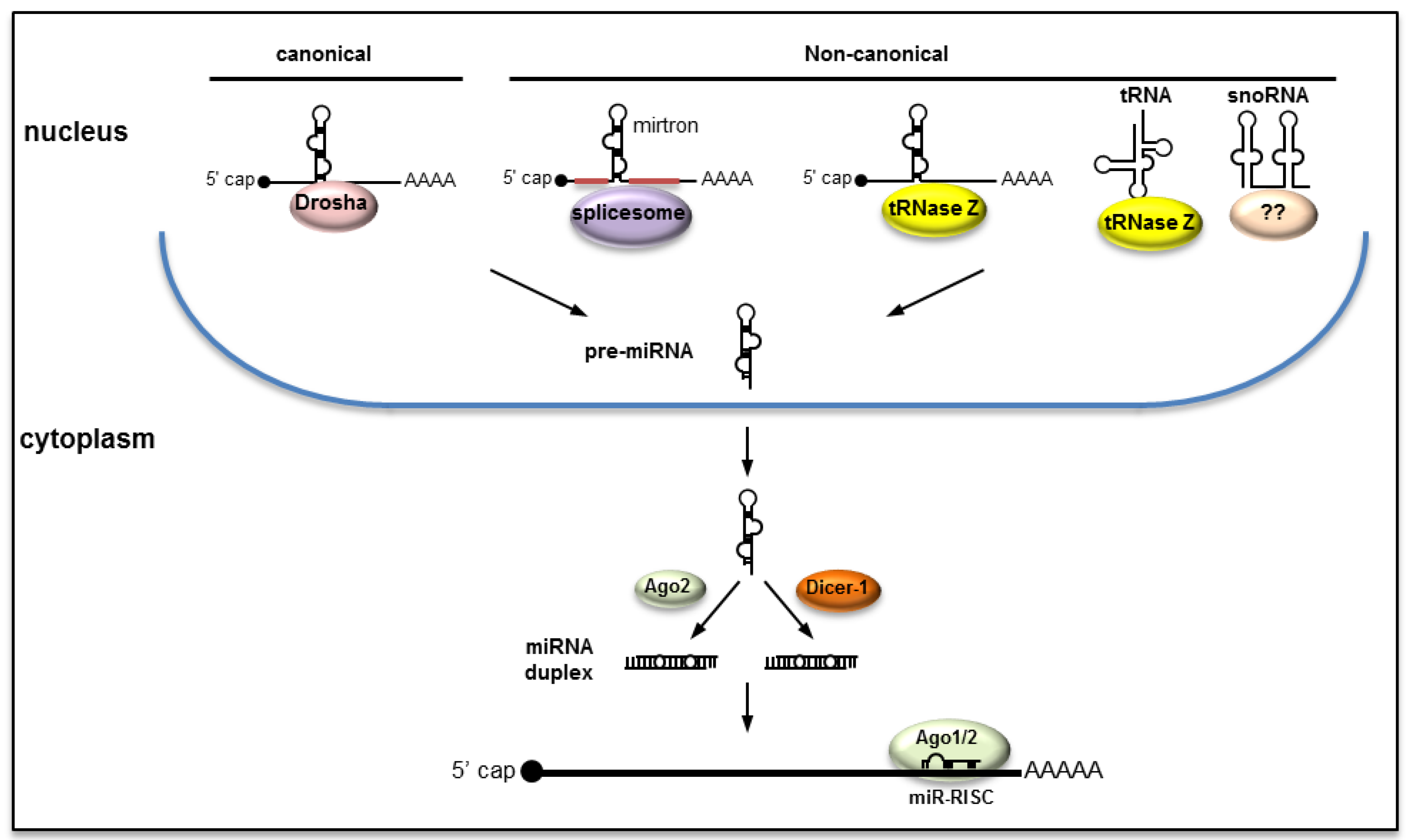
2.2. Non-Canonical Pathways
2.3. Biogenesis of miRNAs from RNA Viruses
3. miRNA-Target Interaction
4. Role of miRNAs in Vector-Arbovirus Interactions
4.1. Effect of Virus Infection on the Host miRNA Profile
4.2. Arbovirus-Encoded miRNAs
| Origin | Name | Function | Insect species/cell type | References |
|---|---|---|---|---|
| Host miRNAs | ||||
| miR-375 | regulates REL1 and cactus genes following blood meal enhancing DENV-2 replication | Ae. aegypti | [28] | |
| miR-2940 | regulates metalloprotease ftsh and Dnmt2 genes; enhances DENV-2 and WNV replication | Ae. aegypti Aag2 and C6/36 cells | [31,43,74] | |
| Viral miRNAs | ||||
| KUN-miR-1 | upregulates cellular GATA4 and facilitates WNV replication | Aag2 and C6/36 cells | [61] | |
| DENV-vsRNA-5 | regulates DENV replication by targeting NS1 sequences | Aag2/RML12 | [67] |
5. Blood Feeding and Its Effects on Arboviruses
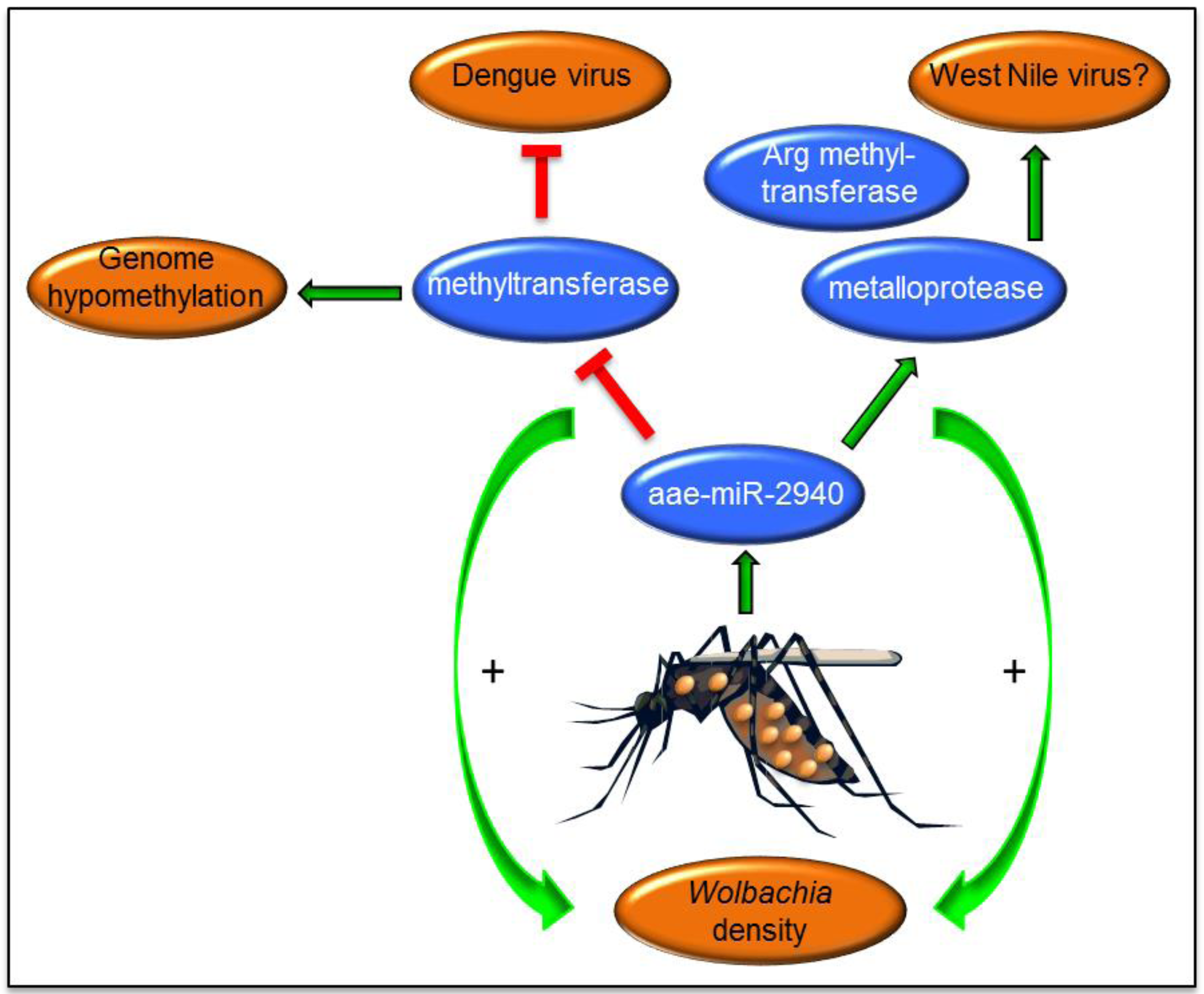
6. Wolbachia-Mediated Alterations of miRNAs and Their Effects on Arbovirus Replication
7. Evolution of miRNAs in Mosquito Vectors
8. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Weaver, S.; Reisen, W. Present and future arboviral threats. Antivir. Res. 2010, 85, 328–345. [Google Scholar] [PubMed]
- Turell, M.J. Arthropod-Related viruses of medical and veterinary importance. In Medical and Veterinary Entomology; Mullen, G., Durden, L., Eds.; Academic Press: Oxford, UK, 2009; pp. 557–564. [Google Scholar]
- Pfeffer, S.; Sewer, A.; Lagos–Quintana, M.; Sheridan, R.; Sander, C.; Grasser, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Ma, Z.; Huo, Y.; Xiao, Z.; Li, Y.; Wang, Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 2011, 17, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Xu, J.; Seitz, H.; Weng, Z.; Zamore, P.D. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 2010, 16, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; O’Neill, S.L.; Asgari, S. Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 2013, 10, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Montañez, R.; Perez, L.; Milan, M.; Belles, X. Regulation of atrophin by both strands of the mir-8 precursor. Insect Biochem. Mol. Biol. 2013, 43, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Smibert, P.; Westholm, J.O.; Jee, D.; Maurin, T.; Lai, E.C. Intertwined pathways for Argonaute-mediated microRNA biogenesis in Drosophila. Nucleic Acids Res. 2014, 42, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Hsu, R.; Melton, C.; Thomas, M.; Ullian, E.M.; Blelloch, R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 2011, 17, 1489–1501. [Google Scholar] [PubMed]
- Bogerd, H.P.; Karnowski, H.W.; Cai, Z.; Shin, J.; Pohlers, M.; Cullen, B.R. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 2010, 37, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.; Glazov, E.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varble, A.; Chua, M.; Perez, J.; Manicassamy, B.; Garcia-Sastre, A.; tenOever, B. Engineered RNA viral synthesis of microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 11519. [Google Scholar] [CrossRef] [PubMed]
- Bennasser, Y.; Le, S.; Yeung, M.; Jeang, K. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 2004, 1, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouha, H.; Thurner, C.; Mandl, C.W. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010, 38, 8328–8337. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.S.; Varble, A.; Pham, A.M.; tenOever, B.R. Noncanonical cytoplasmic processing of viral microRNAs. RNA 2010, 16, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.P.; Langlois, R.A.; Pham, A.M.; tenOever, B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 2012, 18, 1338–1346. [Google Scholar] [PubMed]
- Rosewick, N.; Momont, M.; Durkin, K.; Takeda, H.; Caiment, F.; Cleuter, Y.; Vernin, C.; Mortreux, F.; Wattel, E.; Burny, A.; et al. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 2306–2311. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 23, 215–233. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Nam, J.-W.; Farh, K.K.; Chiang, H.R.; Shkumatava, A.; Bartel, D.P. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol. Cell 2010, 38, 789–802. [Google Scholar] [PubMed]
- Wu, S.; Huang, S.; Ding, J.; Zhao, Y.; Liang, L.; Liu, T.; Zhan, R.; He, X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3’ untranslated region. Oncogene 2010, 29, 2302–2308. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Tirabassi, R.; Meyers, H.; Wu, G.; McWeeney, S.; Hook, L.; Nelson, J.A. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 2010, 6, e1000967. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.J.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-Wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Rajewsky, N. The impact of miRNA target sites in coding sequences and in 3’UTRs. PLoS One 2011, 6, e18067. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Walker, T.; O’Neill, S.; Asgari, S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.G.; Karam, R.; Huang, L.; Bhardwaj, A.; Lou, C.H.; Shum, E.Y.; Song, H.W.; Corbett, M.A.; Gifford, W.D.; Gecz, J.; et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 2011, 42, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Frentiu, F.D.; Moreira, L.A.; O’Neill, S.L.; Asgari, S. Wolbachia utilizes host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. USA 2011, 108, 9250–9255. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5' UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Conrad, K.D.; Giering, F.; Erfurth, C.; Neumann, A.; Fehr, C.; Meister, G.; Niepmann, M. MicroRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS One 2013, 8, e56272. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mead, E.A.; Liang, L.; Tu, Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics 2009, 10, 581. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Hu, W.; Wu, J.; Zheng, P.; Chen, M.; James, A.; Chen, X.; Tu, Z. miRNA genes of an invasive vector mosquito, Aedes albopictus. PLoS One 2013, 8, e67638. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.; Vanlandingham, D.L.; Scholle, F.; Higgs, S.; Cullen, B.R. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Winter, F.; Edaye, S.; Hüttenhofer, A.; Brunel, C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007, 35, 6953–6962. [Google Scholar] [CrossRef] [PubMed]
- Mead, E.A.; Tu, Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics 2008, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.L.; Harrison, T.; Hess, A.M.; Ebel, G.D. MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 2014, 23, 132–139. [Google Scholar]
- Yan, H.; Zhou, Y.; Liu, Y.; Deng, Y.; Chen, X. miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J. Med. Virol. 2014, 86, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Shrinet, J.; Jain, S.; Jain, J.; Bhatnagar, R.; Sunil, S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Negl. Trop. Dis. 2014, 8, e2616. [Google Scholar] [CrossRef] [PubMed]
- Slonchak, A.; Hussain, M.; Torres Morales, S.; Asgari, S.; Khromykh, A.A. Expression of mosquito microRNA aae-miR-2940–5p is down-regulated in response to West Nile virus infection to restrict viral replication. J. Virol. 2014, 88, 8457–8467. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Lu, G.; Torres, S.; Edmonds, J.H.; Kay, B.H.; Khromykh, A.A.; Asgari, S. Effect of Wolbachia on replication of West Nile Virus in mosquito cell line and adult mosquitoes. J. Virol. 2013, 87, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Keene, K.; Foy, B.; Sanchez-Vargas, I.; Beaty, B.; Blair, C.; Olson, K. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Donald, C.L.; Human, S.; Watson, M.; Siu, R.W.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, M.; Arias-Goeta, C.; Martin, E.; O’Hara, Z.; Lulla, A.; Mousson, L.; Rainey, S.; Misbah, S.; Schnettler, E.; Donald, C.; et al. Characterization of Aedes aegypti innate-immune pathways that limit Chikungunya virus replication. Plos Neglect. Trop. Dis. 2014, 8, e2994. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Z.; O’Loughlin, E.; Lee, T.; Houel, S.; O’Carroll, D.; Tarakhovsky, A.; Ahn, N.G.; Yi, R. Quantitative functions of Argonaute proteins in mammalian development. Gene Dev. 2012, 26, 693–704. [Google Scholar] [CrossRef]
- Trobaugh, D.; Gardner, C.; Sun, C.; Haddow, A.; Wang, E.; Chapnik, E.; Mildner, A.; Weaver, S.; Ryman, K.; Klimstra, W. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 2013, 506, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, D.; Jones, I.; Gould, E.; Gritsun, T. Molecular archaeology of Flaviviridae untranslated regions: Duplicated RNA structures in the replication enhancer of flaviviruses and pestiviruses emerged via convergent evolution. PLoS One 2014, 9, e92056. [Google Scholar] [CrossRef] [PubMed]
- Whisnant, A.; Kehl, T.; Bao, Q.; Materniak, M.; Kuzmak, J.; Löchelt, M.; Cullen, B. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J. Virol. 2014, 88, 4679–4686. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Smith, L.P.; Nair, V.; Watson, M. An avian retrovirus uses canonical expression and processing mechanisms to generate viral microRNA. J. Virol. 2014, 88, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.; Skalsky, R.; Kennedy, E.; Furuse, Y.; Whisnant, A.; Flores, O.; Schultz, K.; Putnam, N.; Barrows, N.; Sherry, B.; et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 2014, 88, 8065–8076. [Google Scholar] [CrossRef] [PubMed]
- Acosta, E.; Kumar, A.; Bartenschlager, R. Revisiting dengue virus-host cell interaction: New insights into molecular and cellular virology. Adv. Virus Res. 2013, 88, 1–109. [Google Scholar]
- Roby, J.; Hall, R.A.; Khromykh, A.A. Flavivirus replication and assembly. In Molecular Virology and Control of Flaviviruses; Shi, P.-Y., Ed.; Caister Academic Press: Norfolk, UK, 2012; pp. 21–50. [Google Scholar]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Roby, J.A.; Pijlman, G.P.; Wilusz, J.; Khromykh, A.A. Noncoding subgenomic Flavivirus RNA: multiple Functions in West Nile virus pathogenesis and modulation of host responses. Viruses 2014, 6, 404–427. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; van der Aa, L.; Liu, W.; Palmenberg, A.C.; Hall, R.A.; Khromykh, A.A. A highly structured, nuclease-resistant noncoding RNA produced by flaviviruses is required for pathogenicicty. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Sterken, M.G.; Leung, J.Y.; Metz, S.W.; Geertsema, C.; Goldbach, R.W.; Vlak, J.M.; Kohl, A.; Khromykh, A.A.; Pijlman, G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 2012, 86, 13486–13500. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Torres, S.; Schnettler, E.; Funk, A.; Grundhoff, A.; Pijlman, G.P.; Khromykh, A.A.; Asgari, S. West Nile virus encodes a microRNA-like small RNA in the 3’ untranslated region which upregulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012, 40, 2210–2223. [Google Scholar] [CrossRef] [PubMed]
- Kokoza, V.A.; Martin, D.; Mienaltowski, M.J.; Ahmed, A.; Morton, C.M.; Raikhel, A.S. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene 2001, 274, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Attardo, G.M.; Hansen, I.A.; Raikhel, A.S. GATA factor translation is the final downstream step in the amino acid/target-of-rapamycin-mediated vitellogenin gene expression in the anautogenous mosquito Aedes aegypti. J. Biol. Chem. 2006, 281, 11167–11176. [Google Scholar] [CrossRef] [PubMed]
- Cheon, H.M.; Shin, S.W.; Bian, G.; Park, J.H.; Raikhel, A.S. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J. Biol. Chem. 2006, 281, 8426–8435. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Khromykh, A.A.; Parton, R.G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2007, 2, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [PubMed]
- Hussain, M.; Asgari, S. MicroRNA-Like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc. Natl. Acad. Sci. USA 2014, 111, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Olson, K.E.; Blair, C.D.; Garcia-Blanco, M.A.; Cullen, B.R. A “microRNA-like” small RNA expressed by Dengue virus? Proc. Natl. Acad. Sci. USA 2014, 111, E2359. [Google Scholar] [CrossRef]
- Hussain, M.; Asgari, S. Reply to Skalsky et al.: A microRNA-like small RNA from Dengue virus. Proc. Natl Acad Sci USA 2014, 111, E2360. [Google Scholar]
- Finol, E. Viral small RNA in dengue virus: Are they regulating viral replication beyond serotype 2? Proc. Natl. Acad. Sci. USA 2014, 111, E2915–E2916. [Google Scholar] [CrossRef]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chein, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Taft, R.J.; Asgari, S. An insect virus-encoded microRNA regulates viral replication. J. Virol. 2008, 82, 9164–9170. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Wu, C.P.; Liu, C.Y.Y.; Hsu, P.W.-C.; Wu, E.C.; Chao, Y.C. A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA. Sci. Rep. 2011, 1, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Hussain, M.; O’Neill, S.L.; Asgari, S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase contributing to dengue virus inhibition in Aedes aegypti. Proc. Natl. Acad. Sci. USA 2013, 110, 10276–10281. [Google Scholar] [CrossRef] [PubMed]
- Price, D.; Nagarajan, V.; Churbanov, A.; Houde, P.; Milligan, B.; Drake, L.; Gustafson, J.; Hansen, I. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post- blood meal. PLoS One 2011, 6, e22573. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Radtke, A.; Choi, Y.; Mendes, A.; Valenzuela, J.; Dimopoulos, G. Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. BMC Genomics 2010, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Bryant, B.; Macdonad, W.; Raikhel, A.S. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2010, 107, 22391–22398. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Dunn, W.; Campbell, C.; Olson, K.; Dimon, M.; Marinotti, O.; James, A. RNA-seq analyses of blood-induced changes in gene expression in the mosquito vector species, Aedes aegypti. BMC Genomics 2011, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, Y.; Cao, J.; Zhang, H.; Yu, Y. Distinctive microRNA profiles in the salivary glands of Haemaphysalis longicornis related to tick blood-feeding. Exp. Appl. Acarol. 2013, 59, 339–349. [Google Scholar] [PubMed]
- Xu, J.; Hopkins, K.; Sabin, L.; Yasunaga, A.; Subramanian, H.; Lamborn, I.; Gordesky-Gold, B.; Cherry, S. ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc. Natl. Acad. Sci. USA 2013, 110, 15025–15030. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Dunn, W.; Campbell, C.; Olson, K.; Marinotti, O.; James, A. Strain variation in the transcriptome of the Dengue fever vector, Aedes aegypti. G3 Genes, Genomes, Genetics 2012, 2, 103–114. [Google Scholar]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef] [PubMed]
- Luplertlop, N.; Surasombatpattana, P.; Patramool, S.; Dumas, E.; Wasinpiyamongkol, L.; Saune, L.; Hamel, R.; Bernard, E.; Sereno, D.; Thomas, F.; et al. Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with dengue virus. PLoS Pathog. 2012, 7, e1001252. [Google Scholar] [CrossRef]
- Sim, S.; Dimopoulos, G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One 2010, 5, e10678. [Google Scholar] [CrossRef] [PubMed]
- Fragkoudis, R.; Chi, Y.; Siu, R.W.; Barry, G.; Attarzadeh-Yazdi, G.; Merits, A.; Nash, A.A.; Fazakerley, J.K.; Kohl, A. Semliki Forest virus strongly reduces mosquito host defence signaling. Insect Mol. Biol. 2008, 17, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Sanders, H.; Foy, B.; Evans, A.; Ross, L.; Beaty, B.; Olson, K.; Gill, S. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 218, 215–220. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 2012, 7, e38544. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, A.A.; Turelli, M. Cytoplasmic incompatibility in insects. In Influential Passengers: Inherited Microorganisms and Arthropod Reproduction; O'Neill, S.L., Hoffman, A.A., Werren, J.H., Eds.; Oxford University Press: Oxford, UK, 1997; pp. 42–80. [Google Scholar]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Ferreira, A.; Ashburner, M. The Bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, 2753–2763. [Google Scholar]
- McMeniman, C.J.; Lane, R.V.; Cass, B.N.; Fong, A.W.C.; Sidhu, M.; Wang, Y.-F.; O’Neill, S.L. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 2009, 323, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.J.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with Dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar]
- Bourtzis, K.; Dobson, S.L.; Xi, Z.; Rasgon, J.L.; Calvitti, M.; Moreira, L.A.; Bossin, H.C.; Moretti, R.; Baton, L.A.; Hughes, G.L.; et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014, 132, S150–S163. [Google Scholar] [CrossRef] [PubMed]
- Rainey, S.; Shah, P.; Kohl, A.; Dietrich, I. Understanding the Wolbachia-mediated inhibition of arboviruses in mosquitoes: Progress and challenges. J. Gen. Virol. 2014, 95, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Osborne, S.E.; Leong, Y.; O’Neill, S.L.; Johnson, K.N. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009, 5, e1000656. [Google Scholar] [CrossRef] [PubMed]
- Dodson, B.; Hughes, G.; Paul, O.; Matacchiero, A.; Kramer, L.; Rasgon, J. Wolbachia enhances West Nile virus (WNV) infection in the mosquito Culex tarsalis. PLoS Neglect. Trop. Dis. 2014, 8, e2965. [Google Scholar] [CrossRef]
- Ye, Y.H.; Woolfit, M.; Huttley, G.A.; Rancès, E.; Caragata, E.P.; Popovici, J.; O'Neill, S.L.; McGraw, E.A. Infection with a virulent strain of Wolbachia disrupts genome wide-patterns of cytosine methylation in the mosquito Aedes aegypti. PLoS One 2013, 8, e66482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Hussain, M.; Asgari, S. Regulation of arginine methyltransferase 3 by a Wolbachia-induced microRNA in Aedes aegypti and its effect on Wolbachia and dengue virus replication. Insect Biochem. Mol. Biol. 2014, 53, 81–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayoral, J.G.; Etebari, K.; Hussain, M.; Khromykh, A.A.; Asgari, S. Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS One 2014, 9, e96107. [Google Scholar] [PubMed]
- Lu, J.; Shen, Y.; Wu, Q.; Kumar, S.; He, B.; Shi, S.; Carthew, R.; Wang, S.; Wu, C. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008, 40, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fu, Y.; Kumar, S.; Shen, Y.; Zeng, K.; Xu, A.; Carthew, R.; Wu, C. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol. Biol. Evol. 2008, 25, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, S.; Simmons, M.; Olson, K.; Beaty, B.; Blair, C.; Black, W. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PLoS One 2012, 7, e44198. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Joshi, D.; Dong, Y.; Lu, P.; Zhou, G.; Pan, X.; Xu, Y.; Dimopoulos, G.; Xi, Z. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 2013, 340, 748–751. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Asgari, S. Role of microRNAs in Arbovirus/Vector Interactions. Viruses 2014, 6, 3514-3534. https://doi.org/10.3390/v6093514
Asgari S. Role of microRNAs in Arbovirus/Vector Interactions. Viruses. 2014; 6(9):3514-3534. https://doi.org/10.3390/v6093514
Chicago/Turabian StyleAsgari, Sassan. 2014. "Role of microRNAs in Arbovirus/Vector Interactions" Viruses 6, no. 9: 3514-3534. https://doi.org/10.3390/v6093514




