In the Shadow of Hemagglutinin: A Growing Interest in Influenza Viral Neuraminidase and Its Role as a Vaccine Antigen
Abstract
:1. Influenza Virus: Epidemiology and Basic Virology
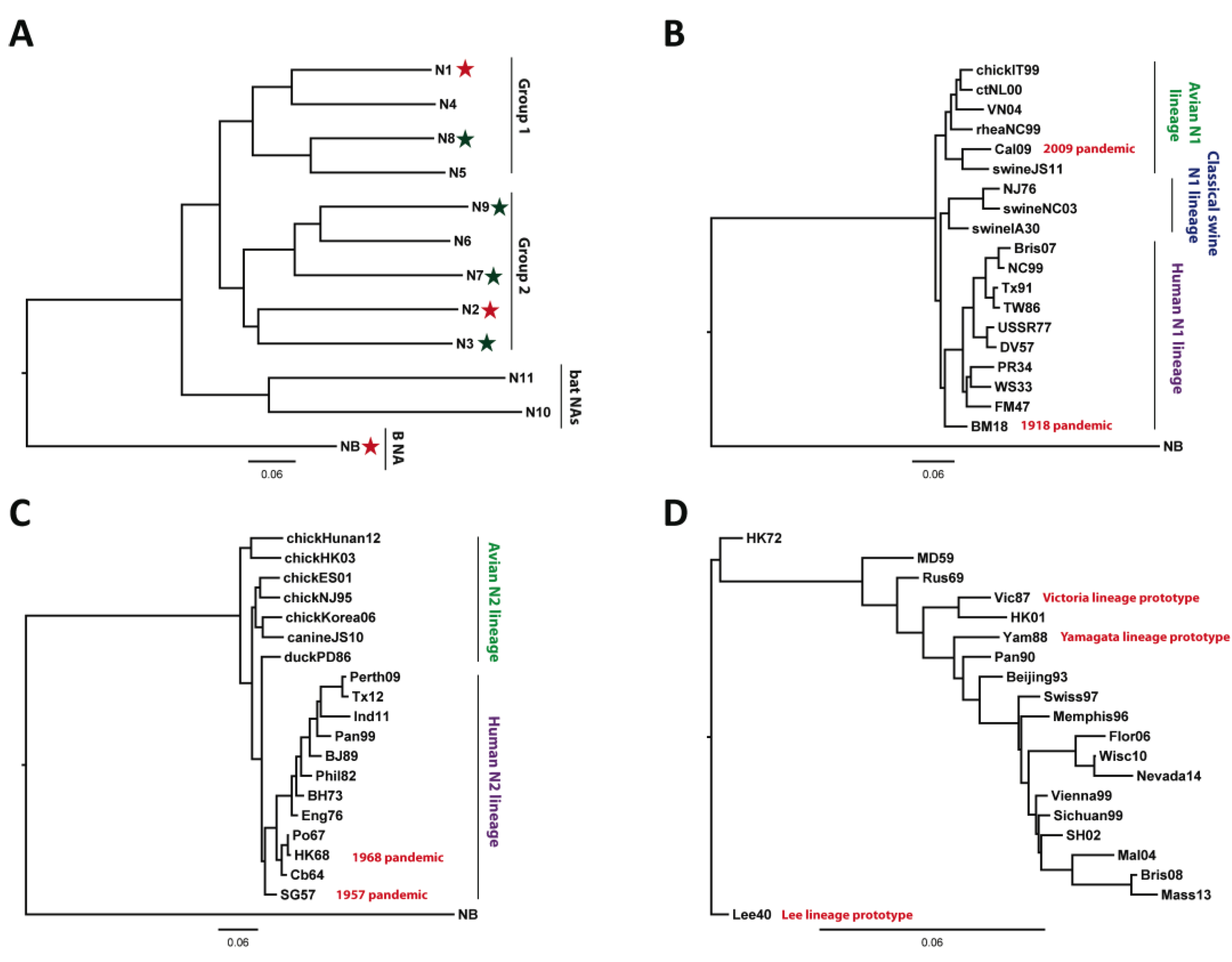
2. Current Therapeutic/Prophylactic Approaches
3. Influenza Neuraminidase: Structure, Function, and Role in Influenza Virus Infection
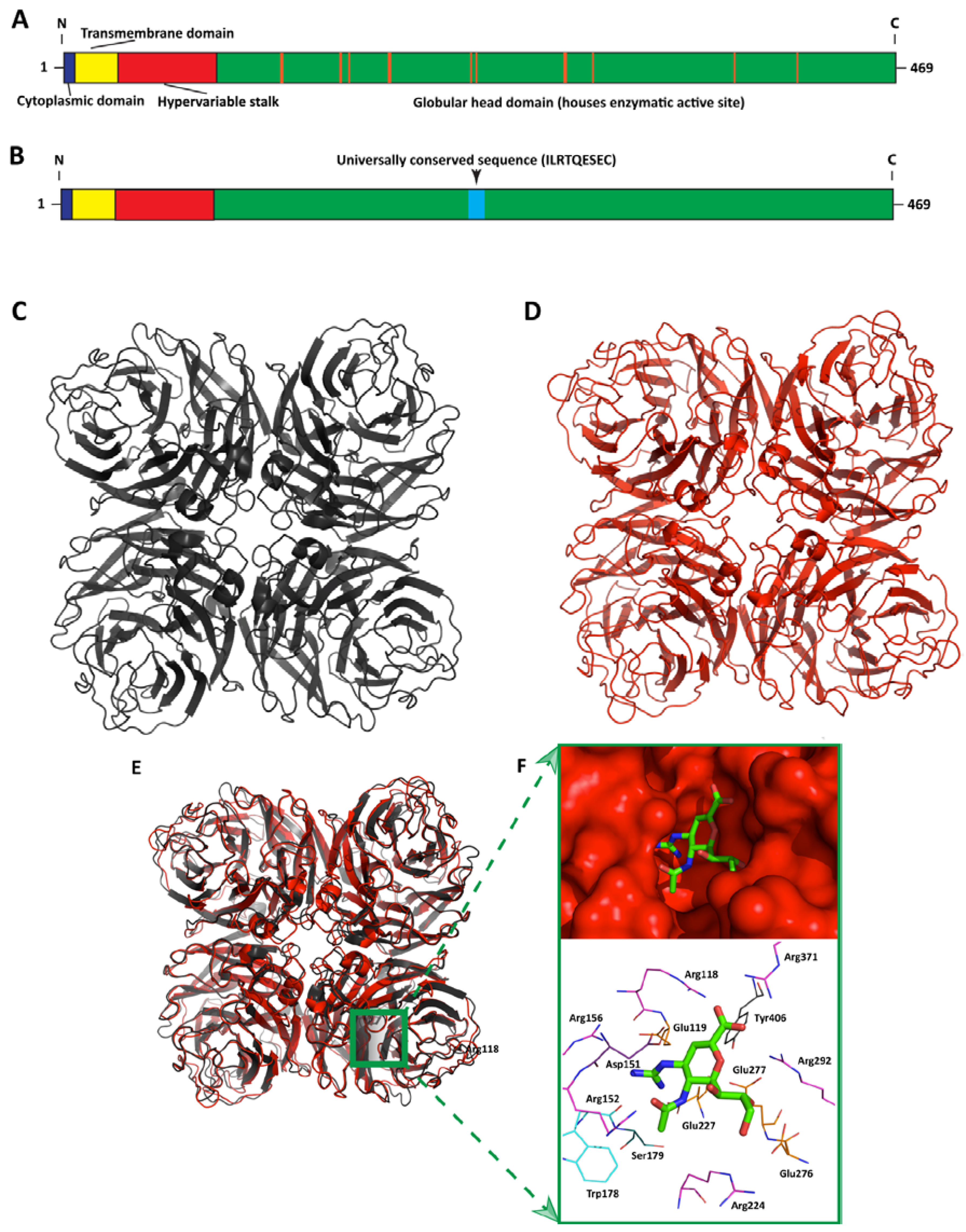
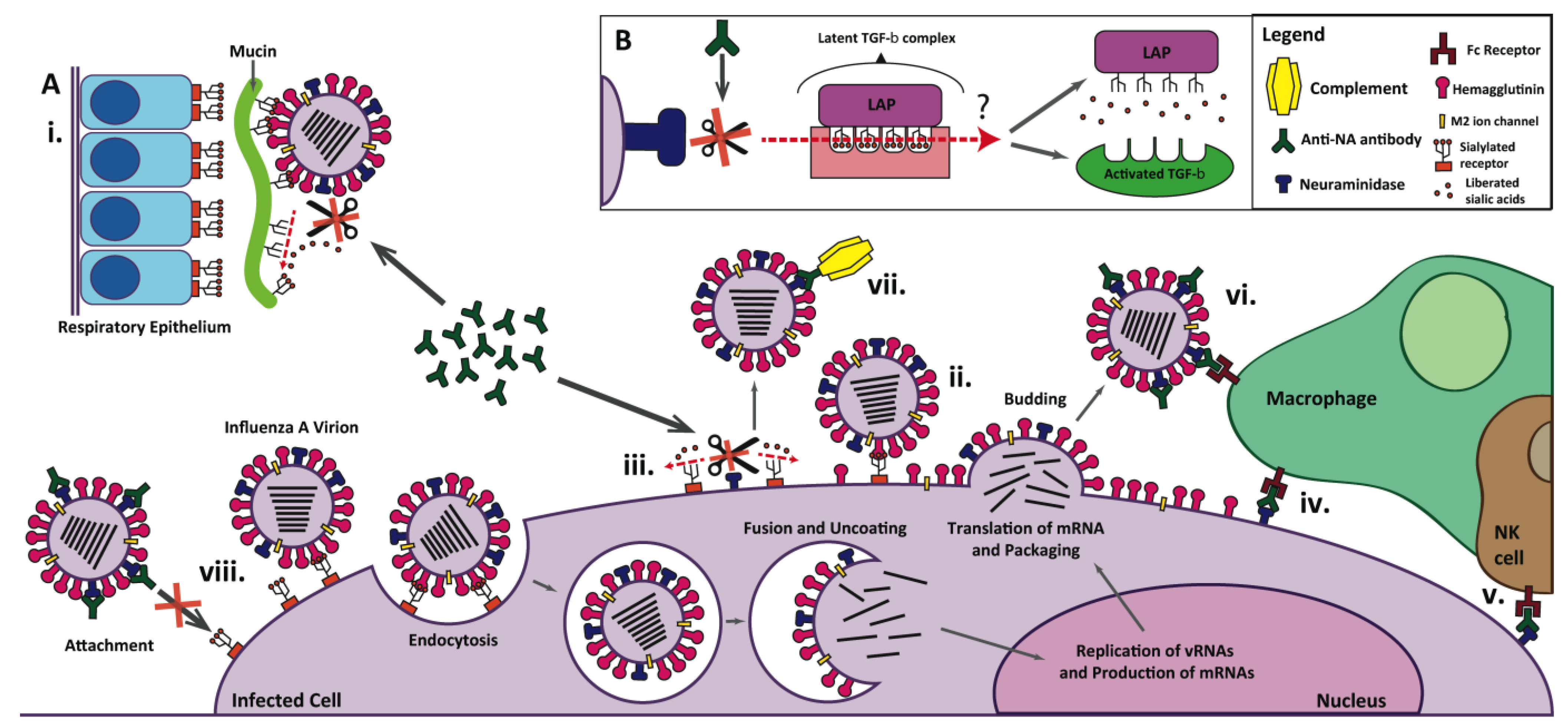

4. Neuraminidase as a Vaccine Antigen
4.1. Immunodominance of HA over NA
4.2. Enhancing the Immunogenicity of NA
4.3. NI Seroconversion Rates in Modern Human Vaccine Trials
| Technology | Model | Main Findings | Ref. |
|---|---|---|---|
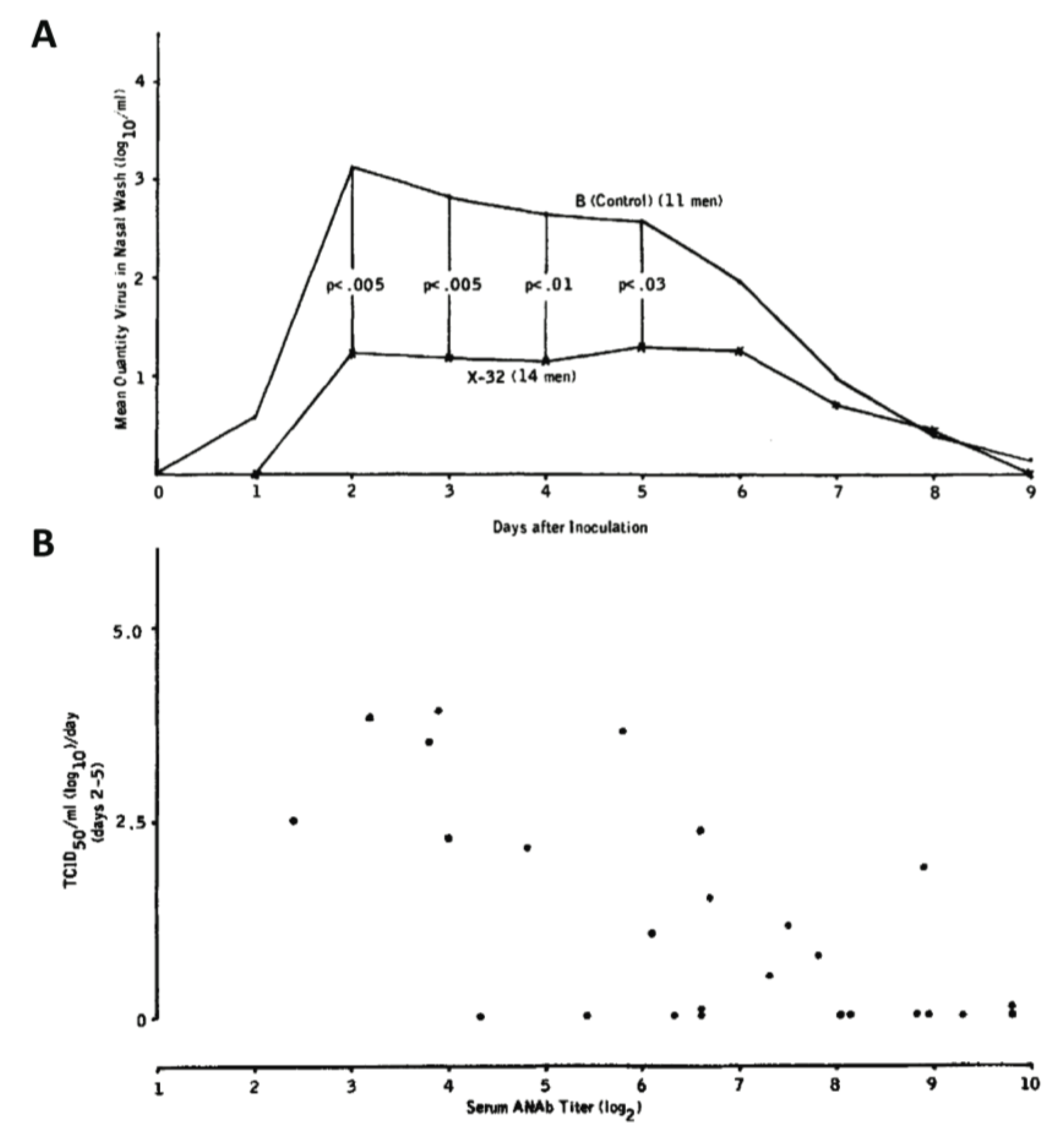
| X-32 (H7N2) reassortant strain vaccine | Humans* | Texan prisoners immunized with an H7N2 reassortant strain displayed significantly lower nasal wash titers when challenged with an infectious dose of circulating H3N2 than those immunized with influenza B virus and the nasal wash titers were inversely proportional to serum anti-NA titer | [89] |
| X-42 (H7N2) reassortant strain vaccine | Humans | School children from Buffalo, NY immunized with an H7N2 reassortant strain displayed a lower attack rate of naturally circulating H3N2 virus than those receiving placebo vaccination | [73] |
| N2 purified from A/Beijing/32/92 virus | Mice, rabbits, and humans | NA was isolated from virus disrupted with detergent and purified (by affinity chromatography using oxamic acid agarose) treated with formalin, and tested for enzymatic activity and immunogenicity in BALB/c mice, rabbits, and humans. The vaccine was shown to be immunogenic and non-toxic, overall | [98,99] |
| N2 from A/Victoria/3/75 expressed in yeast | Mice | Mice were protected against a lethal challenge of influenza virus after immunization with a recombinant head domain of NA, expressed in the yeast species P. pastoris | [100] |
| N2 DNA vaccine | Mice | Mice immunized with NA-expressing plasmid DNA were protected against lethal challenge with homologous and heterologous H3N2 strains, but failed to be protected from H1N1 challenge | [101,102] |
| TIV supplemented with chromatographically purified N1 and N2 | Mice | Mice immunized with TIV supplemented with chromatographically purified N1 and N2 showed a greater reduction in viral lung titers than those receiving TIV alone after challenged with heterotypic strains | [95] |
| Soluble NA produced in a cell-free system | Ferrets | Immunization with soluble tetrameric NA reduces signs of morbidity after influenza virus infection (such as bodyweight loss and lung pathology) upon challenge with a homologous H1N1 strain | [103] |
| A/Vietnam/1203/2004 (H5N1) whole-virus inactivated vaccine produced in Vero cell culture | Humans | Phase I/II clinical trials of individuals vaccinated with H5N1 produced in Vero cell culture displayed an average NI seroconversion rate of 59.9% against N1 | [83] |
| Recombinant, hypoglycosylated NA produced in the yeast species, Pichia pastoris | Mice | Mice immunized with a recombinant, hypoglycosylated form of NA showed higher NI titers than those immunized with an equivalent form of hyperglycosylated NA | [97] |
| H5N1 VLPs (produced by baculovirus infected insect cells) | Mice | Mice immunized with H5N1 VLPs derived from the avian influenza virus strain RG14 H5N1 showed robust NI seroconversion and in-vivo protection against lethal challenge with the matched H5N1 strain and Cal09 H1N1, but were not protected from challenge with PR8 H1N1 | [104] |
| 293T-produced VLPs containing NA and matrix proteins from H1N1 | Mice | Immunization of VLPs protects mice from lethal infection with H5N1 | [105] |
| VLPs containing the N1 and M1 from PR8 (produced by baculovirus-infected insect cells) | Mice | Mice immunized intranasally with VLPs containing N1 and M1 were completely protected against a lethal challenge with the matched, homologous PR8 strain (H1N1) and against a heterosubtypic H3N2 strain (A/Philippines/2/82), although the mice challenged with the H3N2 virus were not protected from weight loss | [106] |
| H7N9 VLPs (produced by baculovirus-infected insect cells) | Humans | Humans immunized with H7N9 (A/Anhui/1/13) VLPs supplemented with ISCOMATRIX adjuvant displayed an average NI seroconversion rate of 97.2% against N9 | [85] |
| Intramuscular immunization with seasonal split TIV | Ferrets | Ferrets that received two intramuscular vaccinations with split TIV vaccines were protected against lethal challenge with H5N1 | [107] |
| Mouse monoclonal antibodies | Mice | Mice prophylactically given one dose of broadly reactive N1 mAbs were protected against lethal challenge with seasonal H1N1, pandemic 2009 H1N1, and H5N1 | [48] |
| Two priming immunizations with A/Vietnam/1203/2004 (H5N1) whole-virus inactivated vaccine produced in Vero cell culture followed by a boost of A/Indonesia/05/2005 | Humans | Test of safety and efficacy of a previously mentioned Vero cell-derived H5N1 vaccine [83] in 675 children aged 6 months–17 years showed no adverse effects and 93.1%–100% of subjects displayed MN titers of 1:20 or greater against the H5N1. NI titers were induced in greater than 90% of subjects. | [84] |
4.4. Historic Evidence of NA-Based Protection in Humans
4.5. Anti-NA Antibodies Affect Viral Evolution
4.6. Future Considerations in the Design of NA-Based Vaccines
| Name | Binding Profile | Organism/Method of Isolation | IC50 Range | Key Findings | Ref. |
|---|---|---|---|---|---|
| 2B9 | N1 NAs from highly pathogenic H5N1 avian influenza viruses as well as N1 NAs from H1N1 strains | Mouse mAb—mice were immunized with recombinant N1 NA from A/Vietnam/1194/2004 and antibody was isolated using hybridoma technology | <1–250 µg/mL (as measured by NI) | 2B9 was shown to enzymatically inhibit NAs from both homologous and heterologous H5N1 strains, as well as some H1N1 strains. Mice passively immunized with 2B9 were 50% protected against lethal challenge with a closely related H5N1 virus | [115] |
| 3A2, 4G2, 1H5, 2D9, and several others | N1 NA from A/Brisbane/59/2007 (seasonal H1N1), Cal09 (pandemic H1N1) and A/Vietnam/1203/2004 (avian H5N1) | Mouse mAb—mice were immunized twice intranasally with A/Brisbane/59/2007 and boosted a third time with purified virus | 7.7 ng/mL–>32,000 ng/mL
(as measured by NI) | Some of the mAbs isolated were broadly reactive against the N1 of seasonal H1N1, pandemic 1918 H1N1, pandemic 2009 H1N1, and H5N1 viruses. A single dose of one mAb, 3A2, was able to fully protect mice against lethal challenge with seasonal and 2009 pandemic H1N1 viruses and resulted in significant protection against wild-type H5N1 | [48] |
| HCA-2 MAb | Universally conserved 9-amino acid long NA sequence, ILRTQESEC | Rabbit mAb—9 amino acid long peptide (ILRTQESEC) was conjugated to a 6-aminocaproic-lysine-lysine-cysteine linker and used to immunize rabbits. Antibodies were produced using hybridoma technology by a commercial contract research organization. | 2.56–18.2 µg/mL (as measured by growth inhibition of various viral strains)
0.95–181.93 µg/mL (as measured by NI of representative NA subtypes) | HCA-2 mAb was shown to inhibit the enzymatic activity of practically all known influenza A NA subtypes (N1-9) as well as those from both the Yamagata and Victoria influenza B virus lineages | [46,47] |
4.7. Evidence of NA-Based Heterologous Protection
4.8. Evidence of NA-Based Heterosubtypic Protection
4.9. Antibodies against H1N1 Protect against H5N1 in Multiple Animal Models
5. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Pappas, G.; Kiriaze, I.J.; Falagas, M. E. Insights into infectious disease in the era of Hippocrates. Int. J. Infect. Dis. 2008, 12, 347–350. [Google Scholar] [CrossRef]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef]
- Wright, P.F.; Neumann, G.; Kawaoka, Y. Orthomyxoviruses. Fields Virol. 2013, 2, 1693–1740. [Google Scholar]
- The World Health Organization. Influenza (Seasonal): Fact Sheet No. 211; The World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Molinari, N.A.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef]
- Shaw, M.; Palese, P. Orthomyxoviridae: The Viruses and Their Replication. Fields Virol. 2013, 2, 1648–1689. [Google Scholar]
- CDC. Types of Influenza Viruses|Seasonal Influenza (Flu); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013. [Google Scholar]
- CDC. Seasonal Influenza (Flu)—Weekly Report: Influenza Summary Update; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013. [Google Scholar]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef]
- Arzey, G.G.; Kirkland, P.D.; Arzey, K.E.; Frost, M.; Maywood, P.; Conaty, S.; Hurt, A.C.; Deng, Y.; Barr, I.; Dwyer, D.E.; et al. Influenza Virus A (H10N7) in Chickens and Poultry Abattoir Workers, Australia. Emerg. Infect. Dis. 2012, 18, 814–816. [Google Scholar] [CrossRef]
- Beare, A.S.; Webster, R.G. Replication of avian influenza viruses in humans. Arch. Virol. 1991, 119, 37–42. [Google Scholar] [CrossRef]
- CDC Update: Influenza Activity—United States and Worldwide, 2002–2003 Season, and Composition of the 2003–2004 Influenza Vaccine. Available online: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5222a2.htm/ (accessed on 6 January 2013).
- Johansson, B.E.; Cox, M.M.J. Influenza viral neuraminidase: The forgotten antigen. Expert Rev. Vaccines 2011, 10, 1683–1695. [Google Scholar] [CrossRef]
- Kayali, G.; Ortiz, E.J.; Chorazy, M.L.; Gray, G.C. Evidence of previous avian influenza infection among US turkey workers. Zoonoses Public Health 2010, 57, 265–272. [Google Scholar]
- Kayali, G.; Barbour, E.; Dbaibo, G.; Tabet, C.; Saade, M.; Shaib, H.A; Debeauchamp, J.; Webby, R.J. Evidence of infection with H4 and H11 avian influenza viruses among Lebanese chicken growers. PLoS One 2011, 6, e26818. [Google Scholar]
- WHO/CDS/CSR/RMD EID Weekly Updates: Emerging and Reemerging Infectious Diseases, Region of the Americas. Available online: http://www1.paho.org/English/AD/DPC/CD/eid-eer-07-may-2004.htm#birdflu/ (accessed on 18 November 2013).
- Yuan, J.; Zhang, L.; Kan, X.; Jiang, L.; Yang, J.; Guo, Z.; Ren, Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013, 57, 1367–1368. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, H.; Gao, R.; Zhang, J.; Wang, D.; Xiong, Y.; Fan, G.; Yang, F.; Li, X.; Zhou, J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: A descriptive study. Lancet 2014, 383, 714–721. [Google Scholar] [CrossRef]
- Pica, N.; Palese, P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu. Rev. Med. 2013, 64, 189–202. [Google Scholar]
- Zachary, K.C. Treatment of Seasonal Influenza in Adults. Available online: http://www.uptodate.com/contents/treatment-of-seasonal-influenza-in-adults?detectedLanguage=en&source=search_result&search=influenza&selectedTitle=1%7E150&provider=noProvider#H13/ (accessed on 18 November 2013).
- Krammer, F.; Grabherr, R. Alternative influenza vaccines made by insect cells. Trends Mol. Med. 2010, 16, 313–320. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Tsai, T.F.; del Giudice, G. New technologies for influenza vaccines. Hum. Vaccines Immunother. 2012, 8, 45–58. [Google Scholar] [CrossRef]
- Quan, F.-S.; Huang, C.; Compans, R.W.; Kang, S.-M. Virus-like particle vaccine induces protective immunity against homologous and heterologous strains of influenza virus. J. Virol. 2007, 81, 3514–3524. [Google Scholar] [CrossRef]
- Klausberger, M.; Wilde, M.; Palmberger, D.; Hai, R.; Albrecht, R.A.; Margine, I.; Hirsh, A.; García-Sastre, A.; Grabherr, R.; Krammer, F. One-shot vaccination with an insect cell-derived low-dose influenza A H7 virus-like particle preparation protects mice against H7N9 challenge. Vaccine 2014, 32, 355–362. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef]
- Gerhard, W.; Yewdell, J.; Frankel, M.E.; Webster, R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 1981, 290, 713–716. [Google Scholar] [CrossRef]
- Hensley, S.E.; Das, S.R.; Bailey, A.L.; Schmidt, L.M.; Hickman, H.D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J.R.; et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 2009, 326, 734–736. [Google Scholar] [CrossRef] [Green Version]
- Marcelin, G.; Sandbulte, M.R.; Webby, R.J. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev. Med. Virol. 2012, 22, 267–279. [Google Scholar] [CrossRef]
- Eichelberger, M.; Golding, H.; Hess, M.; Weir, J.; Subbarao, K.; Luke, C.J.; Friede, M.; Wood, D. FDA/NIH/WHO public workshop on immune correlates of protection against influenza A viruses in support of pandemic vaccine development, Bethesda, Maryland, US, December 10–11, 2007. Vaccine 2008, 26, 4299–4303. [Google Scholar] [CrossRef]
- Bright, R.A.; Neuzil, K.M.; Pervikov, Y.; Palkonyay, L. WHO meeting on the role of neuraminidase in inducing protective immunity against influenza infection, Vilamoura, Portugal, September 14, 2008. Vaccine 2009, 27, 6366–6369. [Google Scholar] [CrossRef]
- Hirst, G.K. The quantitiative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 1942, 75, 49–64. [Google Scholar] [CrossRef]
- Burnet, M. quoted in. Aust. J. Exp. Biol. Med. Sci. 1948, 26, 410. [Google Scholar] [CrossRef]
- Seto, J.T.; Drzeniek, R.; Rott, R. Isolation of a low molecular weight sialidase (neuraminidase) from influenza virus. Biochim. Biophys. Acta Enzymol. Biol. Oxid. 1966, 113, 402–404. [Google Scholar] [CrossRef]
- Varghese, J.N.; Laver, W.G.; Colman, P.M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature 1983, 303, 35–40. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 [angst] resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Palese, P.; Schulman, J.L.; Bodo, G.; Meindl, P. Inhibition of influenza and parainfluenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA). Virology 1974, 59, 490–498. [Google Scholar] [CrossRef]
- Von Itzstein, M.; Wu, W.-Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; Phan, T.V.; Smythe, M.L.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar] [CrossRef]
- Air, G.M. Influenza neuraminidase. Influ. Respir. Viruses 2012, 6, 245–256. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, X.; Dwek, R.A.; Stevens, J.; Wilson, I.A. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 2008, 82, 10493–10501. [Google Scholar] [CrossRef]
- Zhu, X.; McBride, R.; Nycholat, C.M.; Yu, W.; Paulson, J.C.; Wilson, I.A. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors. J. Virol. 2012, 86, 13371–13383. [Google Scholar] [CrossRef]
- Paterson, R.G.; Lamb, R.A. Conversion of a class II integral membrane protein into a soluble and efficiently secreted protein: Multiple intracellular and extracellular oligomeric and conformational forms. J. Cell Biol. 1990, 110, 999–1011. [Google Scholar] [CrossRef]
- Bucher, D.J.; Kilbourne, E.D. A 2 (N2) neuraminidase of the X-7 influenza virus recombinant: Determination of molecular size and subunit composition of the active unit. J. Virol. 1972, 10, 60–66. [Google Scholar]
- Technical Note: R&D Systems Structural Characterization of the 1918 Influenza Virus Neuraminidase. Available online: http://www.rndsystems.com/cb_detail_objectname_cb09i2_technical_notes.aspx/ (accessed on 10 November 2013).
- Sylte, M.J.; Suarez, D.L. Influenza neuraminidase as a vaccine antigen. Curr. Top. Microbiol. Immunol. 2009, 333, 227–241. [Google Scholar]
- Laver, G.; Potter, C.W. Influenza virus surface glycoproteins, haemagglutinin and neuraminidase: A personal account. In Perspectives in Medical Virology; Elsevier: Amsterdam, The Netherlands, 2002; Volume 7, pp. 31–47. [Google Scholar]
- Doyle, T.M.; Hashem, A.M.; Li, C.; van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013, 100, 567–574. [Google Scholar] [CrossRef]
- Doyle, T.M.; Li, C.; Bucher, D.J.; Hashem, A.M.; van Domselaar, G.; Wang, J.; Farnsworth, A.; She, Y.-M.; Cyr, T.; He, R.; et al. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem. Biophys. Res. Commun. 2013, 441, 226–229. [Google Scholar] [CrossRef]
- Wan, H.; Gao, J.; Xu, K.; Chen, H.; Couzens, L.K.; Rivers, K.H.; Easterbrook, J.D.; Yang, K.; Zhong, L.; Rajabi, M.; et al. Molecular basis for broad neuraminidase immunity: Conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J. Virol. 2013, 87, 9290–9300. [Google Scholar] [CrossRef]
- Palese, P.; Tobita, K.; Ueda, M.; Compans, R.W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 1974, 61, 397–410. [Google Scholar] [CrossRef]
- Hooper, K.A.; Bloom, J.D. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J. Virol. 2013. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Matrosovich, T.Y.; Roberts, N.A.; Klenk, H.; Gray, T. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. 2004, 78, 12665–12667. [Google Scholar]
- Cohen, M.; Zhang, X.-Q.; Senaati, H.P.; Chen, H.-W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013, 10, 321. [Google Scholar] [CrossRef]
- Li, M.O.; Wan, Y.Y.; Sanjabi, S.; Robertson, A.-K.L.; Flavell, R.A. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006, 24, 99–146. [Google Scholar]
- Barcellos-Hoff, M.H.; Dix, T.A. Redox-Mediated Activation of Latent Transforming Growth factor-beta1. Mol. Endocrinol. 1996, 10, 1077–1083. [Google Scholar]
- Brown, P.; Wakefield, L.; Levinson, A.; Sporn, M. Physicochemical activation of recombinant latent transforming growth factor-beta’s 1, 2, and 3. Growth Factors 1990, 3, 35–43. [Google Scholar] [CrossRef]
- Gantt, K.R.; Schultz-Cherry, S.; Rodriguez, N.; Jeronimo, S.M.B.; Nascimento, E.T.; Goldman, T.L.; Recker, T.J.; Miller, M.A.; Wilson, M.E. Activation of TGF-beta by Leishmania chagasi: Importance for parasite survival in macrophages. J. Immunol. 2003, 170, 2613–2620. [Google Scholar] [CrossRef]
- Schultz-Cherry, S.; Hinshaw, V.S. Influenza virus neuraminidase activates latent transforming growth factor beta. J. Virol. 1996, 70, 8624–8629. [Google Scholar]
- Carlson, C.M.; Turpin, E.A.; Moser, L.A.; O’Brien, K.B.; Cline, T.D.; Jones, J.C.; Tumpey, T.M.; Katz, J.M.; Kelley, L.A.; Gauldie, J.; et al. Transforming growth factor-β: Activation by neuraminidase and role in highly pathogenic H5N1 influenza pathogenesis. PLoS Pathog. 2010, 6, e1001136. [Google Scholar] [CrossRef]
- Webster, R.G.; Brown, L.E.; Laver, W.G. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 1984, 135, 30–42. [Google Scholar] [CrossRef]
- Hurwitz, J.L.; Hackett, C.J.; Elizabeth, C.; Gerhard, W. Murine Th response to influenza virus: Recognition of hemagglutinin, neuraminidase, matrix, and nucleoproteins. J. Immunol. 1994, 134, 1994–1998. [Google Scholar]
- Herrera, M.T.; Gonzalez, Y.; Juárez, E.; Hernández-Sánchez, F.; Carranza, C.; Sarabia, C.; Guzman-Beltran, S.; Manjarrez, M.E.; Muñoz-Torrico, M.; Garcia-Garcia, L.; et al. Humoral and cellular responses to a non-adjuvanted monovalent H1N1 pandemic influenza vaccine in hospital employees. BMC Infect. Dis. 2013, 13, 544. [Google Scholar] [CrossRef]
- Thomas, P.G.; Keating, R.; Hulse-Post, D.J.; Doherty, P.C. Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 2006, 12, 48–54. [Google Scholar] [CrossRef]
- Johansson, B.E.; Moran, T.M.; Bona, C.A.; Popple, S.W.; Kilbourne, E.D. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. II. Sequential infection of mice simulates human experience. J. Immunol. 1987, 139, 2010–2014. [Google Scholar]
- Kilbourne, E.D. Comparative efficacy of neuraminidase-specific and conventional influenza virus vaccines in induction of antibody to neuraminidase in humans. J. Infect. Dis. 1976, 134, 384–394. [Google Scholar] [CrossRef]
- Powers, D.C.; Kilbourne, E.D.; Johansson, B.E. Neuraminidase-specific antibody responses to inactivated influenza virus vaccine in young and elderly adults. Clin. Diagn. Lab. Immunol. 1996, 3, 511–516. [Google Scholar]
- Couch, R.B.; Atmar, R.L.; Keitel, W.A.; Quarles, J.M.; Wells, J.; Arden, N.; Niño, D. Randomized comparative study of the serum antihemagglutinin and antineuraminidase antibody responses to six licensed trivalent influenza vaccines. Vaccine 2012, 31, 190–195. [Google Scholar]
- Sultana, I.; Yang, K.; Getie-Kebtie, M.; Couzens, L.; Markoff, L.; Alterman, M.; Eichelberger, M.C. Stability of neuraminidase in inactivated influenza vaccines. Vaccine 2014, 32, 2225–2230. [Google Scholar] [CrossRef]
- Akram, A.; Inman, R.D. Immunodominance: A pivotal principle in host response to viral infections. Clin. Immunol. 2012, 143, 99–115. [Google Scholar] [CrossRef]
- Johansson, B.E.; Bucher, D.J.; Kilbourne, E.D. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J. Virol. 1989, 63, 1239–1246. [Google Scholar]
- Lamb, R.A. The influenza virus RNA segments and their encoded proteins. In Genetics of Influenza Viruses; Palese, P., Kingsbury, D.M., Eds.; Springer-Verlag Wein: New York, NY, USA, 1983. [Google Scholar]
- Johansson, B.E.; Moran, T.M.; Kilbourne, E.D. Antigen-presenting B cells and helper T cells cooperatively mediate intravirionic antigenic competition between influenza A virus surface glycoproteins. Proc. Natl. Acad. Sci. USA 1987, 84, 6869–6873. [Google Scholar] [CrossRef]
- Johansson, B.E.; Kilbourne, E.D. Dissociation of influenza virus hemagglutinin and neuraminidase eliminates their intravirionic antigenic competition. J. Virol. 1993, 67, 5721–5723. [Google Scholar]
- Ogra, P.L.; Chow, T.; Beutner, K.R.; Rubi, E.; Strussenberg, J.; DeMello, S.; Rizzone, C. Clinical and immunologic evaluation of neuraminidase-specific influenza A virus vaccine in humans. J. Infect. Dis. 1977, 135, 499–506. [Google Scholar]
- Krammer, F.; Palese, P. Universal influenza virus vaccines: Need for clinical trials. Nat. Immunol. 2014, 15, 3–5. [Google Scholar]
- Margine, I.; Hai, R.; Albrecht, R.A.; Obermoser, G.; Harrod, A.C.; Banchereau, J.; Palucka, K.; García-Sastre, A.; Palese, P.; Treanor, J.J.; et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J. Virol. 2013, 87, 4728–4737. [Google Scholar] [CrossRef]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Petersen, E.; Moran, T.M.; Palese, P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010, 6, e1000796. [Google Scholar] [CrossRef]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Ngai, L.; Ekiert, D.C.; Wilson, I.A.; García-Sastre, A.; Moran, T.M.; Palese, P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 18979–18984. [Google Scholar] [CrossRef]
- Hai, R.; Krammer, F.; Tan, G.S.; Pica, N.; Eggink, D.; Maamary, J.; Margine, I.; Albrecht, R.A.; Palese, P. Influenza viruses expressing chimeric hemagglutinins: Globular head and stalk domains derived from different subtypes. J. Virol. 2012, 86, 5774–5781. [Google Scholar]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef]
- Krammer, F.; Pica, N.; Hai, R.; Tan, G.S.; Palese, P. Hemagglutinin stalk-reactive antibodies are boosted following sequential infection with seasonal and pandemic H1N1 influenza virus in mice. J. Virol. 2012, 86, 10302–10307. [Google Scholar]
- Krammer, F.; Margine, I.; Hai, R.; Flood, A.; Hirsh, A.; Tsvetnitsky, V.; Chen, D.; Palese, P. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J. Virol. 2014, 88, 2340–2343. [Google Scholar]
- Krammer, F.; Hai, R.; Yondola, M.; Tan, G.S.; Leyva-Grado, V.H.; Ryder, A.B.; Miller, M.S.; Rose, J.K.; Palese, P.; García-Sastre, A.; et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 2014, 88, 3432–3442. [Google Scholar] [CrossRef]
- Fritz, R.; Sabarth, N.; Kiermayr, S.; Hohenadl, C.; Howard, M.K.; Ilk, R.; Kistner, O.; Ehrlich, H.J.; Barrett, P.N.; Kreil, T.R. A vero cell-derived whole-virus H5N1 vaccine effectively induces neuraminidase-inhibiting antibodies. J. Infect. Dis. 2012, 205, 28–34. [Google Scholar] [CrossRef]
- Van der Velden, M.V.W.; Fritz, R.; Pöllabauer, E.M.; Portsmouth, D.; Howard, M.K.; Kreil, T.R.; Dvorak, T.; Fritsch, S.; Vesikari, T.; Diez-Domingo, J.; et al. Safety and immunogenicity of a vero cell culture-derived whole-virus influenza A(H5N1) vaccine in a pediatric population. J. Infect. Dis. 2014, 209, 12–23. [Google Scholar] [CrossRef]
- Fries, L.F.; Smith, G.E.; Glenn, G.M. A recombinant viruslike particle influenza A (H7N9) vaccine. N. Engl. J. Med. 2013, 369, 2564–2566. [Google Scholar] [CrossRef]
- Schulman, J.L.; Khakpour, M.; Kilbourne, E. Protective effects of specific immunity to viral neuraminidase on influenza virus infection in mice. J. Virol. 1968, 2, 778–776. [Google Scholar]
- Rott, R.; Becht, H.; Orlich, M. The significance of influenza virus neuraminidase in immunity. J. Gen. Virol. 1974, 22, 35–41. [Google Scholar] [CrossRef]
- Monto, A.S.; Kendal, A.P. Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1973, 1, 623–625. [Google Scholar] [CrossRef]
- Couch, R.B.; Kasel, J.A.; Gerin, J.L.; Schulman, J.L.; Kilbourne, E.D.; Lanier, W.S.; Warden, A.; Taylor, B. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J. Infect. Dis. 1974, 129, 411–420. [Google Scholar] [CrossRef]
- Palese, P.; Wang, T.T. Why do influenza virus subtypes die out? A hypothesis. mBio 2011, 2. [Google Scholar] [CrossRef]
- Abed, Y.; Hardy, I.; Li, Y.; Boivin, G. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J. Med. Virol. 2002, 67, 589–595. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Johansson, B.E.; Grajower, B. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc. Natl. Acad. Sci. USA 1990, 87, 786–790. [Google Scholar] [CrossRef]
- Venkatramani, L.; Bochkareva, E.; Lee, J.T.; Gulati, U.; Graeme Laver, W.; Bochkarev, A.; Air, G.M. An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. J. Mol. Biol. 2006, 356, 651–663. [Google Scholar] [CrossRef]
- Xu, J.; Davis, C.T.; Christman, M.C.; Rivailler, P.; Zhong, H.; Donis, R.O.; Lu, G. Evolutionary history and phylodynamics of influenza A and B neuraminidase (NA) genes inferred from large-scale sequence analyses. PLoS One 2012, 7, e38665. [Google Scholar]
- Johansson, B.E.; Pokorny, B.A.; Tiso, V.A. Supplementation of conventional trivalent influenza vaccine with purified viral N1 and N2 neuraminidases induces a balanced immune response without antigenic competition. Vaccine 2002, 20, 1670–1674. [Google Scholar] [CrossRef]
- Cate, T.R.; Rayford, Y.; Niño, D.; Winokur, P.; Brady, R.; Belshe, R.; Chen, W.; Atmar, R.L.; Couch, R.B. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 2010, 28, 2076–2079. [Google Scholar] [CrossRef]
- Yang, Y.-L.; Chang, S.-H.; Gong, X.; Wu, J.; Liu, B. Expression, purification and characterization of low-glycosylation influenza neuraminidase in α-1,6-mannosyltransferase defective Pichia pastoris. Mol. Biol. Rep. 2012, 39, 857–864. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Couch, R.B.; Kasel, J.A.; Keitel, W.A.; Cate, T.R.; Quarles, J.H.; Grajower, B.; Pokorny, B.A.; Johansson, B.E. Purified influenza A virus N2 neuraminidase vaccine is immunogenic and non-toxic in humans. Vaccine 1995, 13, 1799–1803. [Google Scholar] [CrossRef]
- Hocart, M.; Grajower, B.; Donabedian, A.; Pokorny, B.; Whitaker, C.; Kilbourne, E.D. Preparation and characterization of a purified influenza virus neuraminidase vaccine. Vaccine 1995, 13, 1793–1798. [Google Scholar] [CrossRef]
- Martinet, W.; Saelens, X.; Deroo, T.; Neirynck, S.; Contreras, R.; Min Jou, W.; Fiers, W. Protection of mice against a lethal influenza challenge by immunization with yeast-derived recombinant influenza neuraminidase. Eur. J. Biochem. 1997, 247, 332–338. [Google Scholar]
- Chen, Z.; Matsuo, K.; Asanuma, H.; Takahashi, H.; Iwasaki, T.; Suzuki, Y.; Alzawa, C.; Kurata, T.; Tamura, S. Enhanced protection against a lethal influenza virus challenge by immunization with both hemagglutinin- and neuraminidase-expressing DNAs. Vaccine 1999, 17, 653–659. [Google Scholar] [CrossRef]
- Chen, Z.; Kadowaki, S.; Hagiwara, Y.; Yoshikawa, T.; Matsuo, K.; Kurata, T.; Tamura, S. Cross-protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine 2000, 18, 3214–3222. [Google Scholar] [CrossRef]
- Bosch, B.J.; Bodewes, R.; de Vries, R.P.; Kreijtz, J.H.C.M.; Bartelink, W.; van Amerongen, G.; Rimmelzwaan, G.F.; de Haan, C.A.M.; Osterhaus, A.D.M.E.; Rottier, P.J.M. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J. Virol. 2010, 84, 10366–10374. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Yeh, Y.-C.; Chan, J.-T.; Yang, Y.-C.; Yang, J.-R.; Liu, M.-T.; Wu, H.-S.; Hsiao, P.-W. A VLP vaccine induces broad-spectrum cross-protective antibody immunity against H5N1 and H1N1 subtypes of influenza A virus. PLoS One 2012, 7, e42363. [Google Scholar]
- Easterbrook, J.D.; Schwartzman, L.M.; Gao, J.; Kash, J.C.; Morens, D.M.; Couzens, L.; Wan, H.; Eichelberger, M.C.; Taubenberger, J.K. Protection against a lethal H5N1 influenza challenge by intranasal immunization with virus-like particles containing 2009 pandemic H1N1 neuraminidase in mice. Virology 2012, 432, 39–44. [Google Scholar] [CrossRef]
- Quan, F.-S.; Kim, M.-C.; Lee, B.-J.; Song, J.-M.; Compans, R.W.; Kang, S.-M. Influenza M1 VLPs containing neuraminidase induce heterosubtypic cross-protection. Virology 2012, 430, 127–135. [Google Scholar] [CrossRef]
- Rockman, S.; Brown, L.E.; Barr, I.G.; Gilbertson, B.; Lowther, S.; Kachurin, A.; Kachurina, O.; Klippel, J.; Bodle, J.; Pearse, M.; et al. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J. Virol. 2013, 87, 3053–3061. [Google Scholar] [CrossRef]
- Sylte, M.J.; Hubby, B.; Suarez, D.L. Influenza neuraminidase antibodies provide partial protection for chickens against high pathogenic avian influenza infection. Vaccine 2007, 25, 3763–3772. [Google Scholar] [CrossRef]
- Marcelin, G.; Bland, H.M.; Negovetich, N.J.; Sandbulte, M.R.; Ellebedy, A.H.; Webb, A.D.; Griffin, Y.S.; DeBeauchamp, J.L.; McElhaney, J.E.; Webby, R.J. Inactivated seasonal influenza vaccines increase serum antibodies to the neuraminidase of pandemic influenza A(H1N1) 2009 virus in an age-dependent manner. J. Infect. Dis. 2010, 202, 1634–1638. [Google Scholar] [CrossRef]
- Marcelin, G.; DuBois, R.; Rubrum, A.; Russell, C.J.; McElhaney, J.E.; Webby, R.J. A contributing role for anti-neuraminidase antibodies on immunity to pandemic H1N1 2009 influenza A virus. PLoS One 2011, 6, e26335. [Google Scholar]
- Xie, H.; Li, X.; Gao, J.; Lin, Z.; Jing, X.; Plant, E.; Zoueva, O.; Eichelberger, M.C.; Ye, Z. Revisiting the 1976 “swine flu” vaccine clinical trials: Cross-reactive hemagglutinin and neuraminidase antibodies and their role in protection against the 2009 H1N1 pandemic virus in mice. Clin. Infect. Dis. 2011, 53, 1179–1187. [Google Scholar] [CrossRef]
- Smith, D.J. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef]
- Sandbulte, M.R.; Westgeest, K.B.; Gao, J.; Xu, X.; Klimov, A.I.; Russell, C.A.; Burke, D.F.; Smith, D.J.; Fouchier, R.A.M.; Eichelberger, M.C. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc. Natl. Acad. Sci. USA 2011, 108, 20748–20753. [Google Scholar] [CrossRef]
- Van Reeth, K.; Braeckmans, D.; Cox, E.; van Borm, S.; van den Berg, T.; Goddeeris, B.; de Vleeschauwer, A. Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus. Vaccine 2009, 27, 6330–6339. [Google Scholar] [CrossRef]
- Shoji, Y.; Chichester, J.A.; Palmer, G.A.; Farrance, C.E.; Stevens, R.; Stewart, M.; Goldschmidt, L.; Deyde, V.; Gubareva, L.; Klimov, A.; et al. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum. Vaccines 2011, 7, 199–204. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wohlbold, T.J.; Krammer, F. In the Shadow of Hemagglutinin: A Growing Interest in Influenza Viral Neuraminidase and Its Role as a Vaccine Antigen. Viruses 2014, 6, 2465-2494. https://doi.org/10.3390/v6062465
Wohlbold TJ, Krammer F. In the Shadow of Hemagglutinin: A Growing Interest in Influenza Viral Neuraminidase and Its Role as a Vaccine Antigen. Viruses. 2014; 6(6):2465-2494. https://doi.org/10.3390/v6062465
Chicago/Turabian StyleWohlbold, Teddy John, and Florian Krammer. 2014. "In the Shadow of Hemagglutinin: A Growing Interest in Influenza Viral Neuraminidase and Its Role as a Vaccine Antigen" Viruses 6, no. 6: 2465-2494. https://doi.org/10.3390/v6062465





