Porcine Bocavirus: Achievements in the Past Five Years
Abstract
:1. Introduction
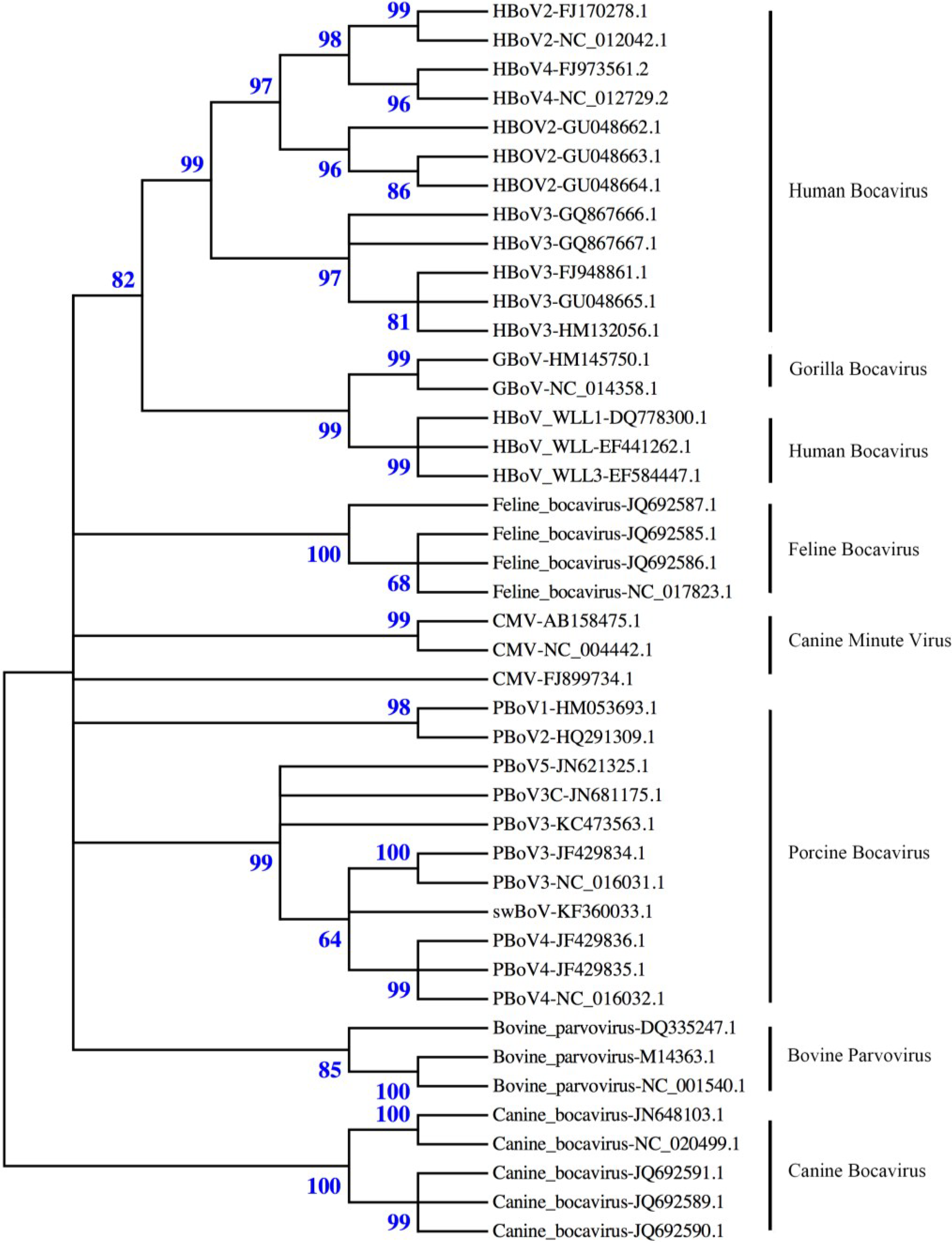
2. Virus Structure
| Published Year | First Author | Country | Name | Accession No. |
|---|---|---|---|---|
| 2009 | Blomstrom, A.L. [19] | Sweden | PBo-likeV | FJ872544 |
| 2010 | Cheng, W.X. [20] | China | PBoV1-CHN | HM053693 |
| PBoV2-CHN | HM053694 | |||
| 6V | HM053672 | |||
| 7V | HM053673 | |||
| 2011 | McKillen, J. [21] | Northern Ireland | PBoV3-UK | JF512472 |
| PBoV4-UK | JF512473 | |||
| 2011 | Lau, S.K. [22] | China | PBoV3-HK | JF429834 |
| PBoV4-HK | JF429835 | |||
| 2012 | Li, B. [24] | China | PBoV5 | JN831651 |
| 2012 | Yang, W.Z. [23] | China | PBoV3C | JN681175 |
| 2014 | Wang, E. [25] | China | swBoV CH437 | KF360033 |

3. Taxonomy and Nomenclature
4. Recombination
5. Detection of Porcine Bocaviruses Using Molecular Virology Techniques
5.1. Cell Culture
5.2. Sequence Detection Assays
5.3. Indirect Immunofluorescence Assay
6. Epidemiology
| Country | Age | Health Condition b | n | Rate (%) | PBoV Type Tested |
|---|---|---|---|---|---|
| Uganda [55] | n/g | n/g | 95 | 2.11% | PBoV G1 |
| Cameroon [56] | piglet | healthy | 50 | 46% | overall |
| USA [57] | mixed | mainly E + R | 385 | 58.7% | overall |
| USA c [43] | n/g | E + R | 203 | 43.3% | overall |
| China [52] | piglet | mainly R + P | 191 | 38.70% | PBoV G1 |
| healthy | 41 | 7.30% | PBoV G1 | ||
| China [20] | piglet | healthy | 397 | 12.59% | PBoV G2 |
| China [28] | pig | healthy | 120 | 39.17% | PBoV G1 |
| China [38] | mixed | healthy | 340 | 63.20% | PBoV G1 |
| 340 | 64.40% | PBoV G2 | |||
| China [39] | pig | clinically sick | 128 | 30.50% | PBoV G1 |
| healthy | 38 | 23.70% | PBoV G1 | ||
| clinically sick | 128 | 21.90% | PBoV G2 | ||
| healthy | 38 | 10.50% | PBoV G2 | ||
| clinically sick | 128 | 38.30% | 6V/7V | ||
| healthy | 38 | 44.70% | 6V/7V | ||
| China [22] | n/g | sick + D | 213 | 18.31% | PBoV G3 |
| healthy | 90 | 16.67% | PBoV G3 | ||
| China [47] | piglet | mainly with PMWS + D | 180 | 56.10% | PBoV G1 |
| healthy | 78 | 16.70% | PBoV G1 | ||
| China [23] | piglet | healthy | 92 | 57.61% | PBoV G2 |
| 92 | 19.60% | PBoV3C | |||
| China [58] | piglet | E | 884 | 31.90% | PBoV G1 |
| healthy | 266 | 26.32% | PBoV G1 | ||
| pig | E | 101 | 27.72% | PBoV G1 | |
| healthy | 58 | 24.14% | PBoV G1 | ||
| China [42] | mixed | E + R + G + D | 403 | 11.41% | overall |
| Korea [45] | mixed | mixed | 920 | 34.9% | overall |
| E + R + G | 351 | 37.8% | overall | ||
| healthy | 679 | 14.9 | overall | ||
| Sweden [19] | piglet | with PMWS | 2 | 100% | PBoV G1 |
| Sweden [54] | piglet | with PMWS | 34 | 88.00% | PBoV G1 |
| healthy | 24 | 46.00% | PBoV G1 | ||
| Northern Ireland [21] | piglet | mainly with PMWS | 369 | 8.70% | PBoV3-UK |
| 369 | 9.50% | PBoV4-UK | |||
| Romania [53] | wild boar | n/g | 470 | 9.14% | PBoV G1 |
| 372 | 17.74% | PBoV G1 | |||
| Hungary [59] | n/g | sick + healthy | 392 | 1.50% | PBoV G1 |
| 392 | 4.80% | PBoV G2 | |||
| 392 | 1.80% | 6V/7V |
7. Pathogenesis
| Group PBoV1(+) Samples with Copathogen b | Listed Virus(+) Samples Coinfected with PBoV c | |
|---|---|---|
| PCV2 (%) | 3.3–83.8 | 37.7 |
| PTTV1 (%) | 73 | n/g |
| PTTV2 (%) | 70.3 | n/g |
| PRRSV (%) | 0–67.6 | 27.3 |
| CSFV (%) | 6.9–34.5 | 20.7 |
| PEDV (%) | 72.6 | 34 |
| PKoV (%) | 72.1 | 32.9 |
| GARV (%) | 9.9 | 41 |
| TGEV (%) | 1 | 66.7 |
8. Conclusions
Authors Contributions
Acknowledgments
Conflicts of Interest
References
- Allander, T.; Tammi, M.T.; Eriksson, M.; Bjerkner, A.; Tiveljung-Lindell, A.; Andersson, B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 2005, 102, 12891–12896. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.M.; Lu, X.; Chittaganpitch, M.; Peret, T.; Fischer, J.; Dowell, S.F.; Anderson, L.J.; Erdman, D.; Olsen, S.J. Human bocavirus: A novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J. Infect. Dis. 2007, 195, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Allander, T.; Jartti, T.; Gupta, S.; Niesters, H.G.; Lehtinen, P.; Osterback, R.; Vuorinen, T.; Waris, M.; Bjerkner, A.; Tiveljung-Lindell, A.; et al. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 2007, 44, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Slikas, E.; Simmonds, P.; Chieochansin, T.; Naeem, A.; Shaukat, S.; Alam, M.M.; Sharif, S.; Angez, M.; Zaidi, S.; et al. A newly identified bocavirus species in human stool. J. Infect. Dis. 2009, 199, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.L.; Higgins, G.D.; Davidson, G.P.; Givney, R.C.; Ratcliff, R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 2009, 5, e1000391. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Simmonds, P.; Slikas, E.; Li, L.; Bodhidatta, L.; Sethabutr, O.; Triki, H.; Bahri, O.; Oderinde, B.S.; Baba, M.M.; et al. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 2010, 201, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Abinanti, F.R.; Warfield, M.S. Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virology 1961, 14, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Inaba, Y.; Kurogi, H.; Omori, T.; Matumoto, M. A new serotype of bovine parvovirus. Jpn. J. Microbiol. 1973, 17, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Allander, T.; Emerson, S.U.; Engle, R.E.; Purcell, R.H.; Bukh, J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA 2001, 98, 11609–11614. [Google Scholar] [CrossRef] [PubMed]
- Pratelli, A.; Moschidou, P. Host range of Canine minute virus in cell culture. J. Vet. Diagn. Investig. 2012, 24, 981–985. [Google Scholar] [CrossRef]
- Decaro, N.; Amorisco, F.; Lenoci, D.; Lovero, A.; Colaianni, M.L.; Losurdo, M.; Desario, C.; Martella, V.; Buonavoglia, C. Molecular characterization of Canine minute virus associated with neonatal mortality in a litter of Jack Russell terrier dogs. J. Vet. Diagn. Investig. 2012, 24, 755–758. [Google Scholar] [CrossRef]
- Kapoor, A.; Mehta, N.; Esper, F.; Poljsak-Prijatelj, M.; Quan, P.L.; Qaisar, N.; Delwart, E.; Lipkin, W.I. Identification and characterization of a new bocavirus species in gorillas. PLoS One 2010, 5, e11948. [Google Scholar] [CrossRef] [PubMed]
- Sharp, C.P.; LeBreton, M.; Kantola, K.; Nana, A.; Diffo, J.D.; Djoko, C.F.; Tamoufe, U.; Kiyang, J.A.; Babila, T.G.; Ngole, E.M.; et al. Widespread infection with homologues of human parvoviruses B19, PARV4, and human bocavirus of chimpanzees and gorillas in the wild. J. Virol. 2010, 84, 10289–10296. [Google Scholar] [CrossRef] [PubMed]
- Babkin, I.V.; Tyumentsev, A.I.; Tikunov, A.Y.; Kurilshikov, A.M.; Ryabchikova, E.I.; Zhirakovskaya, E.V.; Netesov, S.V.; Tikunova, N.V. Evolutionary time-scale of primate bocaviruses. Infect. Genet. Evol. 2013, 14, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Yeung, H.C.; Teng, J.L.; Wu, Y.; Bai, R.; Fan, R.Y.; Chan, K.H.; Yuen, K.Y. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J. Gen. Virol. 2012, 93, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, P.A.; Murphy, B.G. Common and emerging infectious diseases in the animal shelter. Vet. Pathol. 2014, 51, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shan, T.; Wang, C.; Cote, C.; Kolman, J.; Onions, D.; Gulland, F.M.; Delwart, E. The fecal viral flora of California sea lions. J. Virol. 2011, 85, 9909–9917. [Google Scholar] [CrossRef] [PubMed]
- Jartti, T.; Hedman, K.; Jartti, L.; Ruuskanen, O.; Allander, T.; Soderlund-Venermo, M. Human bocavirus-the first 5 years. Rev. Med. Virol. 2012, 22, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Belak, S.; Fossum, C.; McKillen, J.; Allan, G.; Wallgren, P.; Berg, M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009, 146, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.X.; Li, J.S.; Huang, C.P.; Yao, D.P.; Liu, N.; Cui, S.X.; Jin, Y.; Duan, Z.J. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One 2010, 5, e13583. [Google Scholar] [CrossRef] [PubMed]
- McKillen, J.; McNeilly, F.; Duffy, C.; McMenamy, M.; McNair, I.; Hjertner, B.; Millar, A.; McKay, K.; Lagan, P.; Adair, B.; et al. Isolation in cell cultures and initial characterisation of two novel bocavirus species from swine in Northern Ireland. Vet. Microbiol. 2011, 152, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Yip, C.C.; Li, K.S.; Fu, C.T.; Huang, Y.; Chan, K.H.; Yuen, K.Y. Co-existence of multiple strains of two novel porcine bocaviruses in the same pig, a previously undescribed phenomenon in members of the family Parvoviridae, and evidence for inter- and intra-host genetic diversity and recombination. J. Gen. Virol. 2011, 92, 2047–2059. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Yu, J.M.; Li, J.S.; Cheng, W.X.; Huang, C.P.; Duan, Z.J. Genome characterization of a novel porcine bocavirus. Arch. Virol. 2012, 157, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Ma, J.; Xiao, S.; Fang, L.; Zeng, S.; Wen, L.; Zhang, X.; Ni, Y.; Guo, R.; Yu, Z.; et al. Complete genome sequence of a novel species of Porcine Bocavirus, PBoV5. J. Virol. 2012, 86, 1286–1287. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Liu, W.; Yang, B.; Liu, J.; Ma, X.; Lan, X. Complete sequence and phylogenetic analysis of a porcine bocavirus strain swBoV CH437. Virus Genes 2014 2014, 48, 387–390. [Google Scholar] [CrossRef]
- Tijssen, P.; Agbandje-McKenna, M.; Almendral, J.M.; Bergoin, M.; Flegel, T.W.; Hedman, K.; Kleinschmidt, J.; Li, Y.; Pintel, D.J.; Tattersall, P. Family Parvoviridae. In Virus Taxonomy. 9th report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: London, UK, 2012; pp. 405–425. [Google Scholar]
- Zeng, S.; Wang, D.; Fang, L.; Ma, J.; Song, T.; Zhang, R.; Chen, H.; Xiao, S. Complete coding sequences and phylogenetic analysis of porcine bocavirus. J. Gen. Virol. 2011, 92, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, A.Y.; Cheng, F.; Guan, W.; Johnson, F.B.; Qiu, J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 2009, 83, 3956–3967. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.; Cotmore, S.F.; Tattersall, P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 1995, 69, 5422–5430. [Google Scholar] [PubMed]
- Zhang, Z.; Zheng, Z.; Luo, H.; Meng, J.; Li, H.; Li, Q.; Zhang, X.; Ke, X.; Bai, B.; Mao, P.; et al. Human bocavirus NP1 inhibits IFN-beta production by blocking association of IFN regulatory factor 3 with IFNB promoter. J. Immunol. 2012, 189, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Huang, C.P.; Duan, Z.J. Identification and characterization of porcine bocavirus episomes. Bing Du Xue Bao 2012, 28, 418–423. (In Chinese) [Google Scholar] [PubMed]
- Lusebrink, J.; Schildgen, V.; Tillmann, R.L.; Wittleben, F.; Bohmer, A.; Muller, A.; Schildgen, O. Detection of head-to-tail DNA sequences of human bocavirus in clinical samples. PLoS One 2011, 6, e19457. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, P.; Ward, D.C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 1976, 263, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Hirt, B. Molecular biology of autonomous parvoviruses. Contrib. Microbiol. 2000, 4, 163–177. [Google Scholar] [PubMed]
- Cotmore, S.F.; Tattersall, P. Genome packaging sense is controlled by the efficiency of the nick site in the right-end replication origin of parvoviruses minute virus of mice and LuIII. J. Virol. 2005, 79, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Hornig, M.; Asokan, A.; Williams, B.; Henriquez, J.A.; Lipkin, W.I. Bocavirus episome in infected human tissue contains non-identical termini. PLoS One 2011, 6, e21362. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Lan, D.; Li, L.; Wang, C.; Cui, L.; Zhang, W.; Hua, X.; Zhu, C.; Zhao, W.; Delwart, E. Genomic characterization and high prevalence of bocaviruses in swine. PLoS One 2011, 6, e17292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Huang, L.; Liu, Y.J.; Lin, T.; Sun, C.Q.; Deng, Y.; Wei, Z.Z.; Cheung, A.K.; Long, J.X.; Yuan, S.S. Porcine bocaviruses: Genetic analysis and prevalence in Chinese swine population. Epidemiol. Infect. 2011, 139, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs. Infect. Genet. Evol. 2013, 16, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Y.; Sun, D.; Xia, Y.; Huang, J.; Guo, L. Detection and genetic analysis of porcine bocavirus in different swine herds in North Central China. Sci. World J. 2014, 2014, e947084. [Google Scholar]
- Huang, J.; Mor, S.K.; Erber, J.; Voss, E.; Goyal, S.M. Detection and characterization of porcine bocavirus in the United States. Arch. Virol. 2014, 159, 1797–1801. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wang, X.; Ni, B.; Shen, H.; Wang, H.; Zhang, X.; Chen, S.; Shao, S.; Zhang, W. Recombination analysis based on the complete genome of bocavirus. Virol. J. 2011, 8, e182. [Google Scholar] [CrossRef]
- Choi, M.G.; Park, S.J.; Nguyen, V.G.; Chung, H.C.; Kim, A.R.; Park, B.K. Molecular detection and genetic analysis of porcine bocavirus in Korean domestic swine herds. Arch. Virol. 2013, 159, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Yue, C.; Wei, Z.Z.; Ran, D.L.; Long, J.X.; Lin, T.; Deng, Y.; Sun, L.C.; Huang, L.; Yuan, S.S. Development and application of a PCR assay for detection of porcine bocavirus. Chin. J. Anim. Infect. Dis. 2010, 2, 14–17. (In Chinese) [Google Scholar]
- Li, B.; Xiao, S.; Ma, J.; Liu, Y.; Mao, L.; Wen, L.; Mao, A.; Zhang, X.; Ni, Y.; Guo, R.; et al. Development of a novel TaqMan-based real-time PCR assay for the detection of porcine boca-like virus (Pbo-likeV). Virol. J. 2011, 8, e357. [Google Scholar] [CrossRef]
- Li, B.; Ma, J.J.; Xiao, S.B.; Zhang, X.H.; Wen, L.B.; Mao, L.; Ni, Y.X.; Guo, R.L.; Zhou, J.M.; Lv, L.X.; et al. Development of a loop-mediated isothermal amplification method for rapid detection of porcine boca-like virus. J. Virol. Methods 2012, 179, 390–395. [Google Scholar] [CrossRef] [PubMed]
- McNair, I.; McNeilly, F.; Duffy, C.; McKillen, J.; McMenamy, M.; Welsh, M.; Allan, G. Production, characterisation and applications of monoclonal antibodies to two novel porcine bocaviruses from swine in Northern Ireland. Arch. Virol. 2011, 156, 2157–2162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, C.; Gao, M.; He, X.; Diao, Y.; Goyal, S.M.; Mor, S.K.; Huang, J. Evolutionary, epidemiological, demographical, and geographical dissection of porcine bocavirus in China and America. Virus Res. 2014, 3, e217. [Google Scholar]
- Cheung, A.K.; Wu, G.; Wang, D.; Bayles, D.O.; Lager, K.M.; Vincent, A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010, 155, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Yue, C.; Wei, Z.; Long, J.; Ran, D.; Lin, T.; Deng, Y.; Huang, L.; Sun, L.; Zheng, H.; et al. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch. Virol. 2010, 155, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Csagola, A.; Lorincz, M.; Tombacz, K.; Kiss, T.; Spinu, M.; Tuboly, T. Genetic detection and analysis of porcine bocavirus type 1 (PoBoV1) in European wild boar (Sus scrofa). Virus Genes 2011, 43, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Belak, S.; Fossum, C.; Fuxler, L.; Wallgren, P.; Berg, M. Studies of porcine circovirus type 2, porcine boca-like virus and torque teno virus indicate the presence of multiple viral infections in postweaning multisystemic wasting syndrome pigs. Virus Res. 2010, 152, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Stahl, K.; Okurut, A.R.; Masembe, C.; Berg, M. Genetic characterisation of a porcine bocavirus detected in domestic pigs in Uganda. Virus Genes 2013, 47, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Ndze, V.N.; Cadar, D.; Csagola, A.; Kisfali, P.; Kovacs, E.; Farkas, S.; Ngu, A.F.; Esona, M.D.; Dan, A.; Tuboly, T.; et al. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect. Genet. Evol. 2013, 17, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Xiao, C.T.; Yin, S.H.; Gerber, P.F.; Halbur, P.G.; Opriessnig, T. High prevalence and genetic diversity of porcine bocaviruses in pigs in the USA, and identification of multiple novel porcine bocaviruses. J. Gen. Virol. 2014, 95, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013, 158, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Csagola, A.; Lorincz, M.; Cadar, D.; Tombacz, K.; Biksi, I.; Tuboly, T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012, 157, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, Y.; Jiang, J.; Yang, Y.; Liu, K.; Li, Y. High prevelance of human parvovirus infection in patients with malignant tumors. Oncol. Lett. 2012, 3, 635–640. [Google Scholar] [PubMed]
- Schildgen, V.; Malecki, M.; Tillmann, R.L.; Brockmann, M.; Schildgen, O. The Human Bocavirus Is Associated with Some Lung and Colorectal Cancers and Persists in Solid Tumors. PLoS One 2013, 8, e68020. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Sun, H.; Wang, Y. Porcine Bocavirus: Achievements in the Past Five Years. Viruses 2014, 6, 4946-4960. https://doi.org/10.3390/v6124946
Zhou F, Sun H, Wang Y. Porcine Bocavirus: Achievements in the Past Five Years. Viruses. 2014; 6(12):4946-4960. https://doi.org/10.3390/v6124946
Chicago/Turabian StyleZhou, Feng, Haoting Sun, and Yuyan Wang. 2014. "Porcine Bocavirus: Achievements in the Past Five Years" Viruses 6, no. 12: 4946-4960. https://doi.org/10.3390/v6124946




