In between: Gypsy in Drosophila melanogaster Reveals New Insights into Endogenous Retrovirus Evolution
Abstract
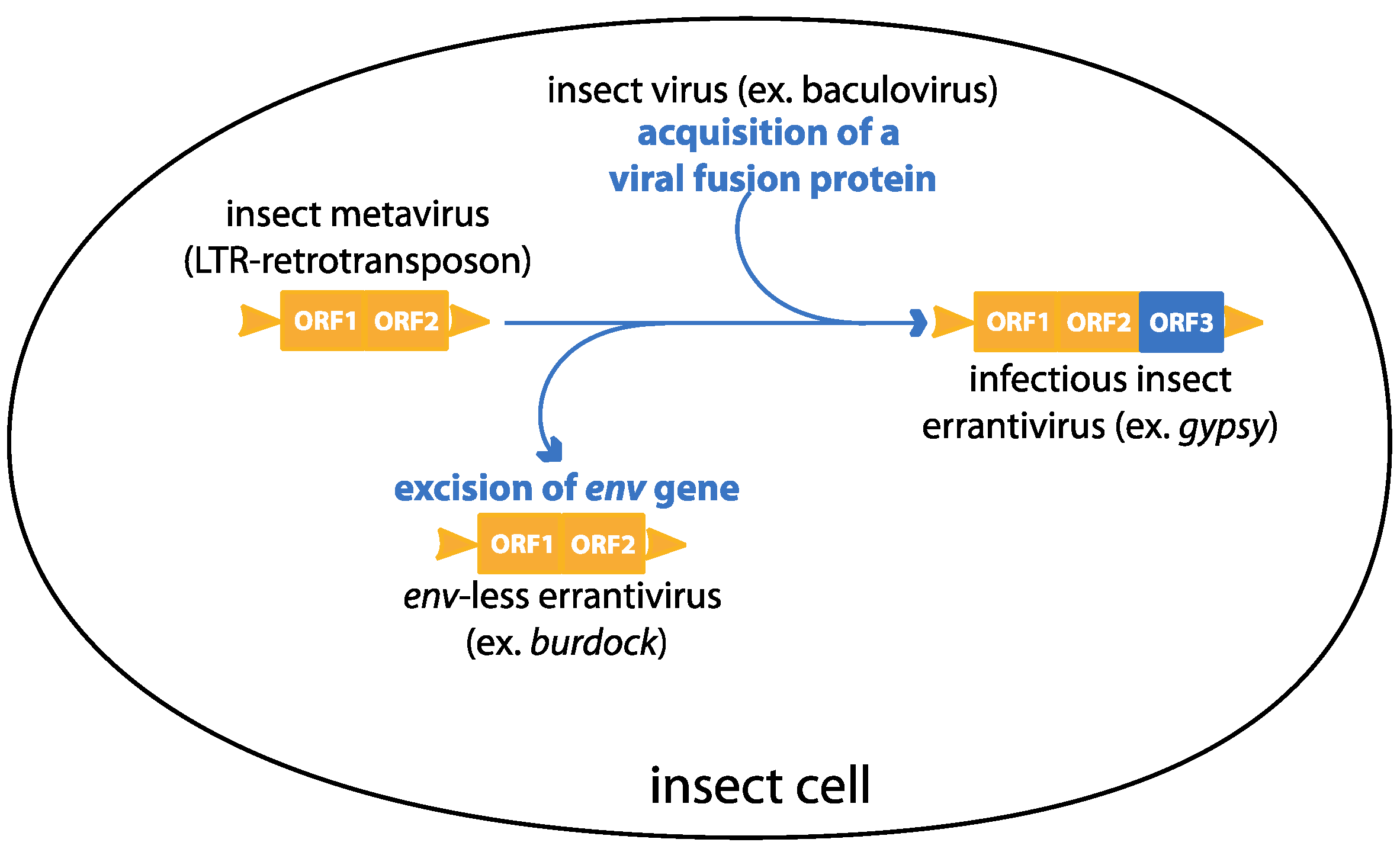
:1. Introduction
2. Gypsy: An Errantivirus of Drosophila melanogaster
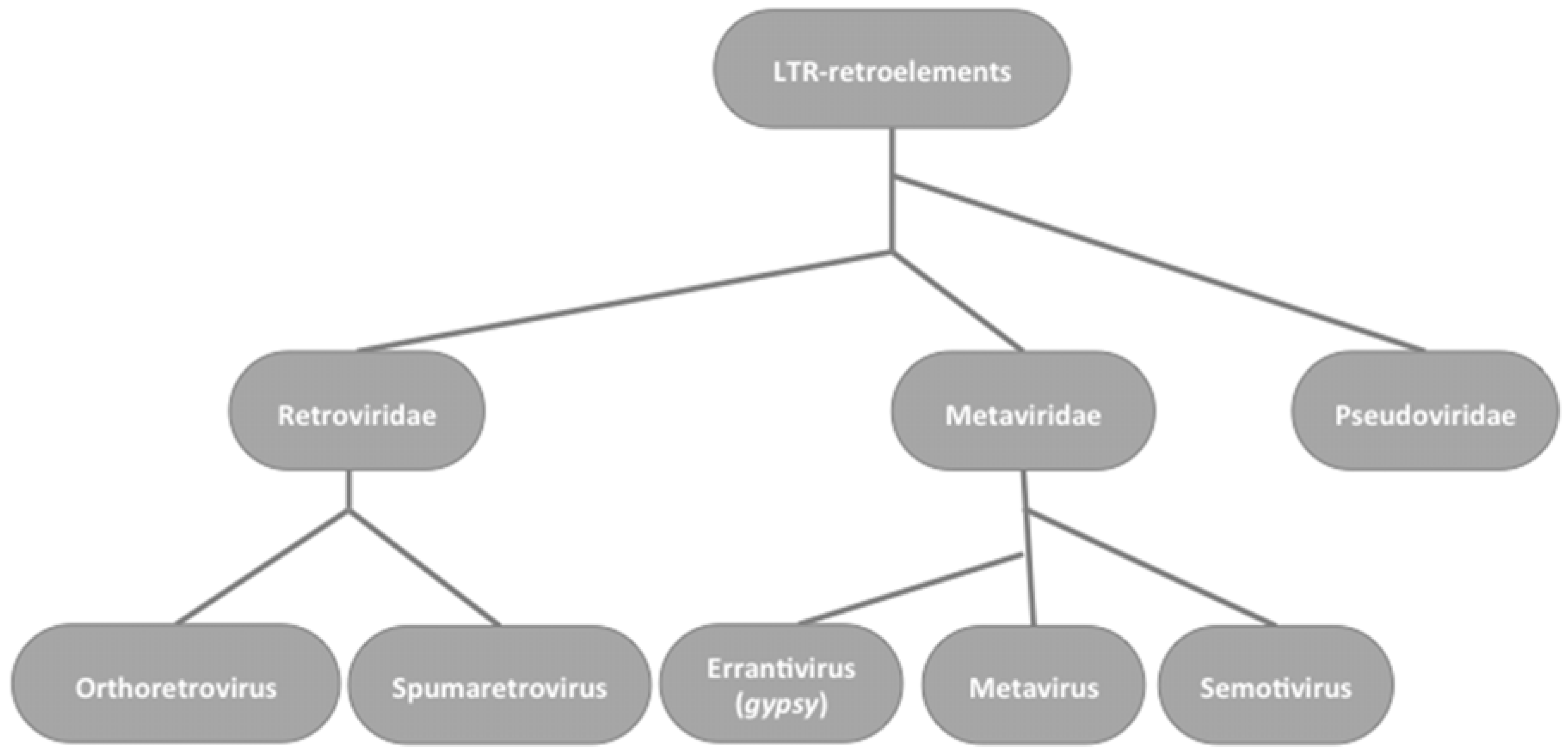
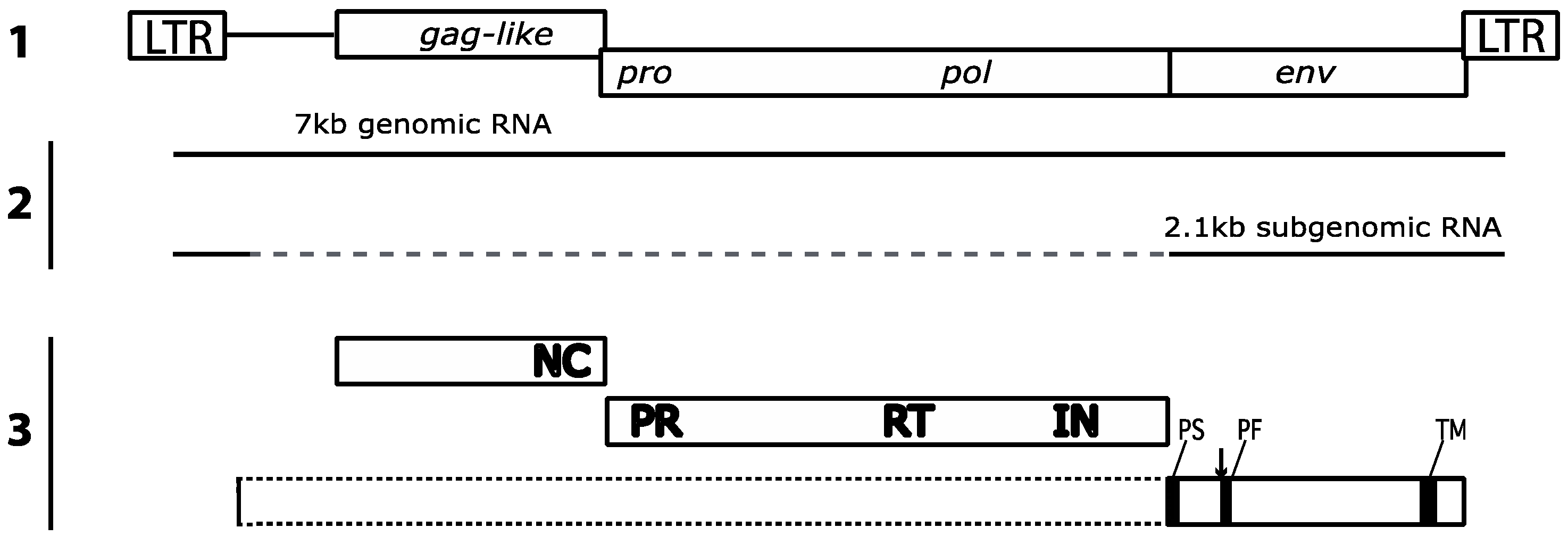
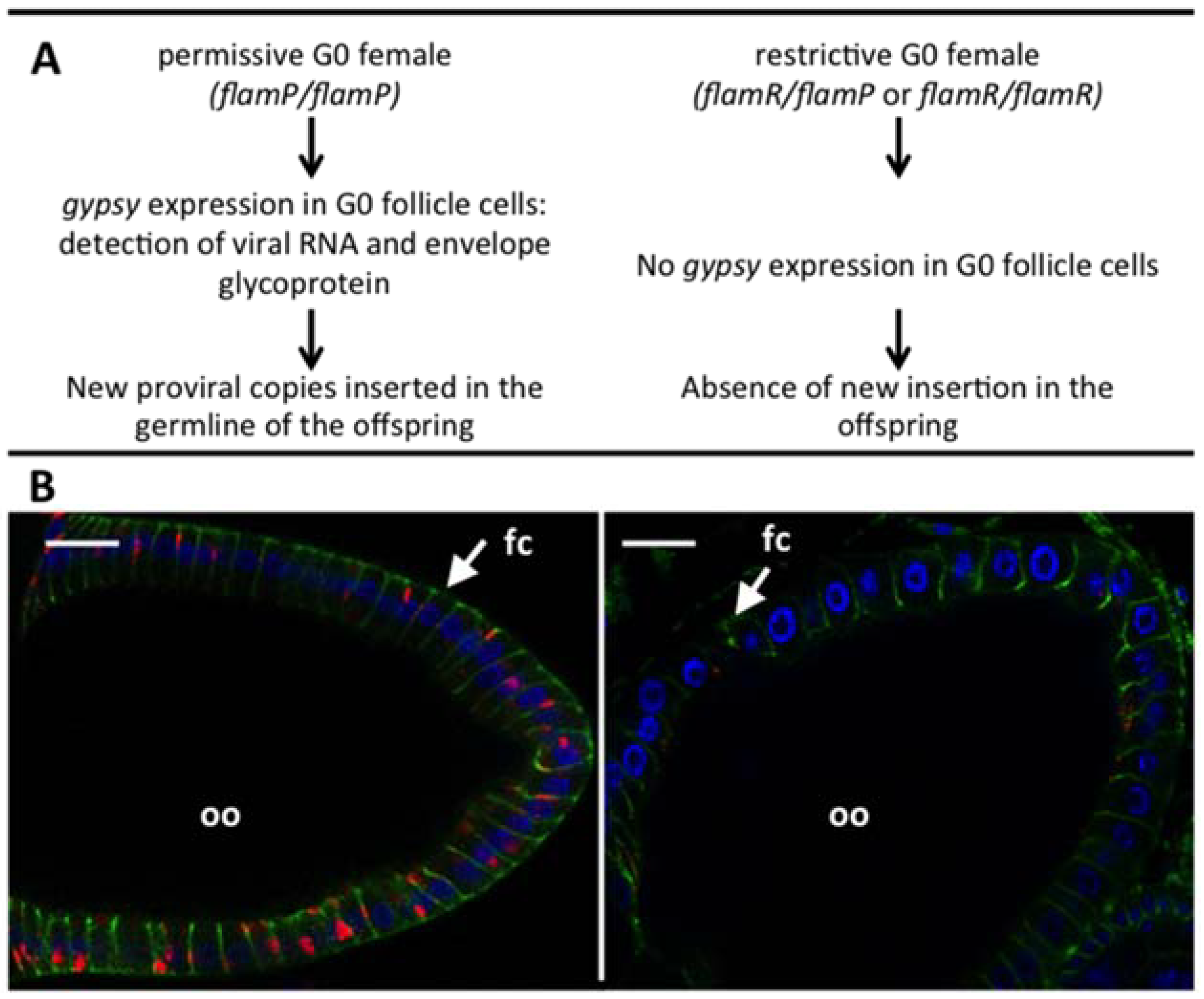
2.1. Gypsy Regulation by Flamenco
| Errantivirus | FlyBase ID |
|---|---|
| 17.6 | FBte0000109 |
| 297 | FBte0000675 |
| gtwin | FBte0001062 |
| gypsy | FBte0000021 |
| gypsy4 | FBte0000688 |
| gypsy5 | FBte0000308 |
| gypsy6 | FBte0001175 |
| Idefix | FBte0000104 |
| Quasimodo | FBte0000640 |
| rover | FBte0000692 |
| springer | FBte0000333 |
| ZAM | FBte0000217 |
2.2. The Genetic “Music” of flamenco
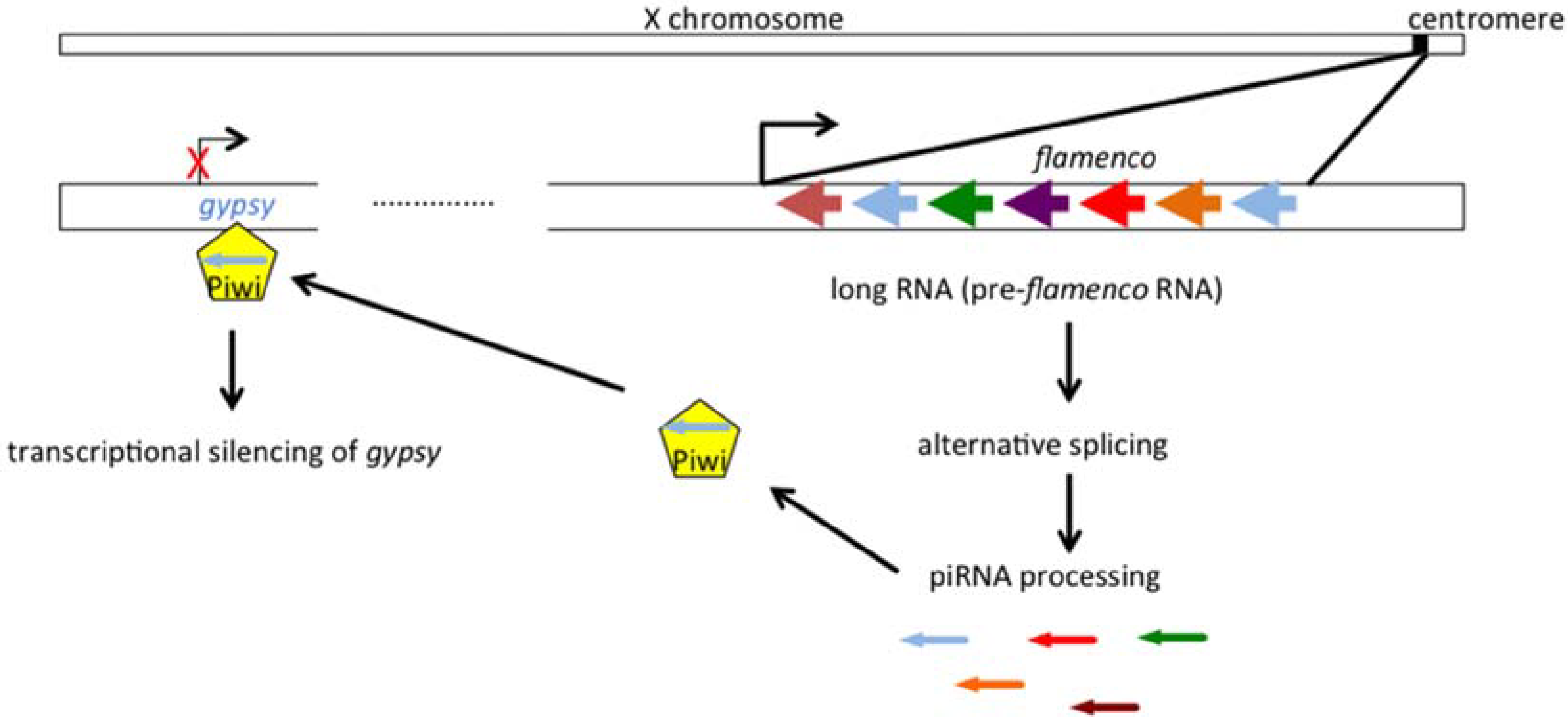
3. Gypsy: An Endogenous Retrovirus with Infectious Properties

4. Wolbachia Influences Gypsy Maternal Transmission
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Katzourakis, A.; Gifford, R.J. Endogenous viral elements in animal genomes. PLOS Genet 2010, 6, e1001191. [Google Scholar] [CrossRef] [PubMed]
- Blikstad, V.; Benachenhou, F.; Sperber, G.O.; Blomberg, J. Evolution of human endogenous retroviral sequences: A conceptual account. Cell. Mol. Life Sci. 2008, 65, 3348–3365. [Google Scholar] [CrossRef] [PubMed]
- Dewannieux, M.; Harper, F.; Richaud, A.; Letzelter, C.; Ribet, D.; Pierron, G.; Heidmann, T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006, 16, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Mallet, F.; Bouton, O.; Prudhomme, S.; Cheynet, V.; Oriol, G.; Bonnaud, B.; Lucotte, G.; Duret, L.; Mandrand, B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. USA 2004, 101, 1731–1736. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.E.; Meers, J.; Young, P.R. Retroviral invasion of the koala genome. Nature 2006, 442, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Gabus, C.; Ivanyi-Nagy, R.; Depollier, J.; Bucheton, A.; Pelisson, A.; Darlix, J.-L. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006, 34, 5764–5777. [Google Scholar] [CrossRef] [PubMed]
- Delelis, O.; Lehmann-Che, J.; Saïb, A. Foamy viruses—A world apart. Curr. Opin. Microbiol. 2004, 7, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S. Positive selection of Iris, a retroviral envelope–derived host gene in Drosophila melanogaster. PLOS Genet. 2005, 1, e44. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, L.N.; Kuzmin, I.V.; Makhnovskii, P.A.; Kim, A.I. Domesticated retroviral GAG gene in Drosophila: New functions for an old gene. Virology 2014, 450–451, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Ronfort, C.; de Breyne, S.; Sandrin, V.; Darlix, J.-L.; Ohlmann, T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. RNA 2004, 10, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.K.; Corces, V.G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992, 6, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Lambertsson, A.; Andersson, S.; Johansson, T. Cloning and characterization of variable-sized gypsy mobile elements in Drosophila melanogaster. Plasmid 1989, 22, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Biémont, C.; Lemeunier, F.; Garcia Guerreiro, M.P.; Brookfield, J.F.; Gautier, C.; Aulard, S.; Pasyukova, E.G. Population dynamics of the copia, mdg1, mdg3, gypsy, and P transposable elements in a natural population of Drosophila melanogaster. Genet. Res. 1994, 63, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Mével-Ninio, M.; Terracol, R.; Kafatos, F.C. The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 1991, 10, 2259–2266. [Google Scholar] [PubMed]
- Prud’homme, N.; Gans, M.; Masson, M.; Terzian, C.; Bucheton, A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 1995, 139, 697–711. [Google Scholar] [PubMed]
- Chalvet, F.; Teysset, L.; Terzian, C.; Prud’homme, N.; Santamaria, P.; Bucheton, A.; Pélisson, A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999, 18, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, P.; Desset, S.; Giorgi, F.; Taddei, A.R.; Fausto, A.M.; Mazzini, M.; Dastugue, B.; Vaury, C. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 2000, 74, 10658–10669. [Google Scholar] [CrossRef] [PubMed]
- Brasset, E.; Taddei, A.R.; Arnaud, F.; Faye, B.; Fausto, A.M.; Mazzini, M.; Giorgi, F.; Vaury, C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 2006, 3, e25. [Google Scholar] [CrossRef] [Green Version]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Goriaux, C.; Desset, S.; Renaud, Y.; Vaury, C.; Brasset, E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014, 15, 411–418. [Google Scholar] [CrossRef]
- Murota, Y.; Ishizu, H.; Nakagawa, S.; Iwasaki, Y.W.; Shibata, S.; Kamatani, M.K.; Saito, K.; Okano, H.; Siomi, H.; Siomi, M.C. Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 2014, 8, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, D.; Senti, K.-A.; Subramanian, S.; Sachidanandam, R.; Brennecke, J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 2012, 47, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Bourc’his, D.; Voinnet, O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 2010, 330, 617–622. [Google Scholar] [CrossRef]
- Sienski, G.; Dönertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Elgin, S.C.R. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA 2011, 108, 21164–21169. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Tóth, K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [PubMed]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Zamore, P.D. piRNAs. Curr. Biol. 2014, 24, R730–R733. [Google Scholar] [CrossRef] [PubMed]
- Zanni, V.; Eymery, A.; Coiffet, M.; Zytnicki, M.; Luyten, I.; Quesneville, H.; Vaury, C.; Jensen, S. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc. Natl. Acad. Sci. USA 2013, 110, 19842–19847. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Terzian, C.; Santamaria, P.; Pelisson, A.; Purd’homme, N.; Bucheton, A. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1994, 91, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Song, S.U.; Gerasimova, T.; Kurkulos, M.; Boeke, J.D.; Corces, V.G. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994, 8, 2046–2057. [Google Scholar] [CrossRef] [PubMed]
- Pelisson, A.; Song, S.U.; Prud’homme, N.; Smith, P.A.; Bucheton, A.; Corces, V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994, 13, 4401–4411. [Google Scholar] [PubMed]
- Rohrmann, G.F.; Karplus, P.A. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol. Biol. 2001, 1, e1. [Google Scholar] [CrossRef]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that baculovirus GP64 superfamily proteins are class III penetrenes. Virol. J. 2008, 5, e28. [Google Scholar] [CrossRef]
- Gallaher, W.R.; DiSimone, C.; Buchmeier, M.J. The viral transmembrane superfamily: Possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 2001, 1, e1. [Google Scholar] [CrossRef]
- Misseri, Y.; Labesse, G.; Bucheton, A.; Terzian, C. Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol. 2003, 11, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Ferree, P.M.; Frydman, H.M.; Li, J.M.; Cao, J.; Wieschaus, E.; Sullivan, W. Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLOS Pathog. 2005, 1, e14. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702–702. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e2. [Google Scholar] [CrossRef] [PubMed]
- Riegler, M.; Sidhu, M.; Miller, W.J.; O’Neill, S.L. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 2005, 15, 1428–1433. [Google Scholar] [CrossRef]
- Touret, F.; Guiguen, F.; Terzian, C. Wolbachia influences the maternal transmission of the gypsy endogenous retrovirus in Drosophila melanogaster. MBio 2014, 5, e01529-14. [Google Scholar] [CrossRef] [PubMed]
- Terzian, C.; Pélisson, A.; Bucheton, A. Evolution and phylogeny of insect endogenous retroviruses. BMC Evol. Biol. 2001, 1, e3. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touret, F.; Guiguen, F.; Greenland, T.; Terzian, C. In between: Gypsy in Drosophila melanogaster Reveals New Insights into Endogenous Retrovirus Evolution. Viruses 2014, 6, 4914-4925. https://doi.org/10.3390/v6124914
Touret F, Guiguen F, Greenland T, Terzian C. In between: Gypsy in Drosophila melanogaster Reveals New Insights into Endogenous Retrovirus Evolution. Viruses. 2014; 6(12):4914-4925. https://doi.org/10.3390/v6124914
Chicago/Turabian StyleTouret, Franck, François Guiguen, Timothy Greenland, and Christophe Terzian. 2014. "In between: Gypsy in Drosophila melanogaster Reveals New Insights into Endogenous Retrovirus Evolution" Viruses 6, no. 12: 4914-4925. https://doi.org/10.3390/v6124914




