Arboviral Bottlenecks and Challenges to Maintaining Diversity and Fitness during Mosquito Transmission
Abstract
:1. Introduction
1.1. Arboviruses
1.2. Bottlenecks
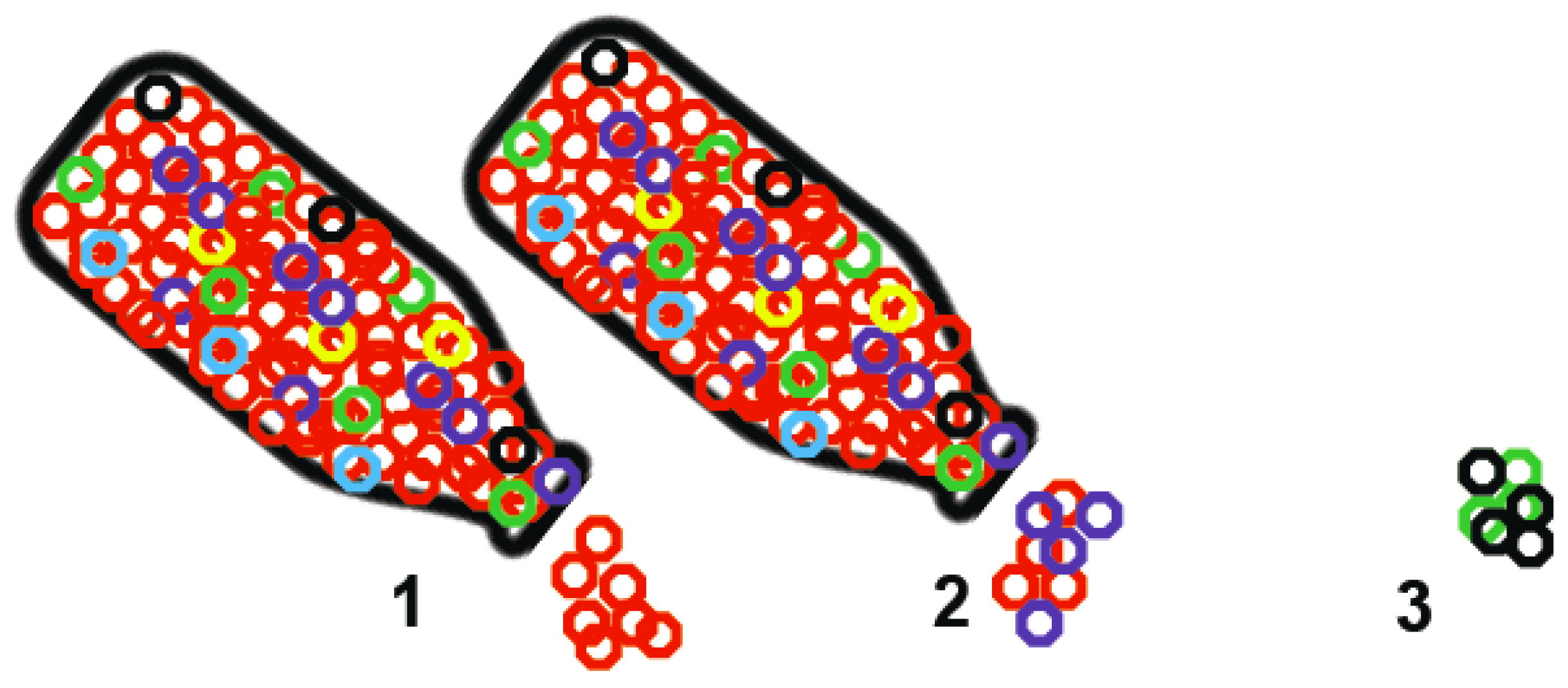
2. Transmission Cycles and Viral Diversity
3. Evolutionary Pressures on Arboviruses and Fitness Potentials
4. Case Studies
4.1. West Nile Transmission and Bottlenecks
4.2. Venezuelan Equine Encephalitis Virus

4.3. Chikungunya Virus
5. Conclusions
Author Contributions
Conflicts of Interest
References and Notes
- Calisher, C.H. Antigenic classification and taxonomy of flaviviruses (family Flaviviridae) emphasizing a universal system for the taxonomy of viruses causing tick-borne encephalitis. Acta Virol. 1988, 32, 469–478. [Google Scholar] [PubMed]
- Karabatsos, N. International Catalogue of Arboviruses, 3rd ed.; American Society of Tropical Medicine and Hygiene: San Antonio, TX, USA, 1985. [Google Scholar]
- Calisher, C.H.; Karabatsos, N. Arbovirus serogroups: Definition and geographic distribution. In The Arboviruses: Epidemiology and Ecology, Vol. I; Monath, T.P., Ed.; CRC Press: Boca Raton, FL, USA, 1988; pp. 19–57. [Google Scholar]
- Coffey, L.L.; Forrester, N.; Tsetsarkin, K.; Vasilakis, N.; Weaver, S.C. Factors shaping the adaptive landscape for arboviruses: Implications for the emergence of disease. Future Microbiol. 2013, 8, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Roossinck, M.J. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 2004, 78, 10582–10587. [Google Scholar] [CrossRef] [PubMed]
- Hawks, J.; Hunley, K.; Lee, S.-H.; Wolpoff, M. Population bottlenecks and pleistocene human evolution. Mol. Biol. Evol. 1999, 17, 2–22. [Google Scholar] [CrossRef]
- Surridge, A.; Bell, D.J.; Ibrahim, K.M.; Hewitt, G.M. Population structure and genetic variation of European wild rabbits (Oryctolagus cuniculus) in East Anglia. Heredity 1999, 82, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Forrester, N.L.; Guerbois, M.; Seymour, R.; Spatt, H.; Weaver, S.C. Vector-borne transmissions imposes a severe bottleneck on an RNA virus population. PLoS Pathog. 2012, 8, e1002897. [Google Scholar] [CrossRef] [PubMed]
- Sacristan, S.; Malpica, J.M.; Fraile, A.; Garcia-Arenal, F. Estimation of population bottlenecks during systemic movement of Tobacco Mosaic virus in Tobacco plants. J. Virol. 2003, 77, 9906–9911. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Maynard-Smith, J. The Evolution of Sex; Cambridge University Press: Cambridge, UK, 1976. [Google Scholar]
- Duate, E.; Clarke, D.; Moya, A.; Domingo, E.; Holland, J. Rapid fitness losses in mammalian RNA clones due to Muller's Ratchet. Proc. Natl. Acad. Sci. 1992, 89, 6015–6019. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Gonzalez-Candelas, F.; Novella, I.S.; Duate, E.A.; Clarke, D.K.; Domingo, E.; Holland, J.J.; Moya, A. Evolution of fitness in experimental populations of vesicular stomatitis virus. Genetics 1996, 142, 673–679. [Google Scholar] [PubMed]
- Escarmis, C.; Davila, M.; Charpentier, N.; Bracho, A.; Moya, A.; Domingo, E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J. Mol. Biol. 1996, 264, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Escarmis, C.; Davila, M.; Domingo, E. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller’s ratchet. J. Mol. Biol. 1999, 285, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Yuste, E.; Lopez-Galindez, C.; Domingo, E. Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type 1. J. Virol. 2000, 74, 9546–9552. [Google Scholar] [CrossRef] [PubMed]
- Yuste, E.; Sanchez-Palomino, S.; Casado, C.; Domingo, E.; Lopez-Galindez, C. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J. Virol. 1999, 73, 2745–2751. [Google Scholar] [PubMed]
- Escarmis, C.; Lazaro, E.; Arias, A.; Domingo, E. Repeated bottleneck transfers can lead to non-cytocidal forms of a cytopathic virus: Implication for viral extinction. J. Mol. Biol. 2008, 376, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Brault, A.C.; Kang, W.; Holland, J.J. Genetic and fitness changes accompanying adaptation of an arbovirus to Vertebrate and Invertebrate cells. J. Virol. 1999, 73, 4316–4326. [Google Scholar] [PubMed]
- Domingo, E.; Escarmis, C.; Lazaro, E.; Manrubia, S.C. Quasispecies dynamics and RNA virus extinction. Virus Res. 2005, 107, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Foy, B.D.; Myles, K.M.; Pierro, D.J.; Sanchez-Vargas, I.; Uhlirova, M.; Jindra, M.; Beaty, B.J.; Olson, K.E. Develoment of a new Sindbis virus transducing system and its characterization in three Culicine mosquitoes and two Lepidopteran species. Insect Mol. Biol. 2004, 13, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.F.; Myles, K.M.; Seabaugh, R.C.; Higgs, S.; Carlson, J.O.; Beaty, B.J. Development of a Sindbis virus expression system that efficiently expresses green fluorescent protein in midguts of Aedes aegypti following per os infection. Insect Mol. Biol. 2000, 9, 57–65. [Google Scholar] [CrossRef]
- Scholle, F.; Girard, Y.A.; Zhao, Q.; Higgs, S.; Mason, P.W. Trans-packaged West Nile virus-like particles: Infectious properties in vitro and in infected mosquito vectors. J. Virol. 2004, 78, 11605–11614. [Google Scholar] [CrossRef]
- Smith, D.; Adams, A.P.; Kenney, J.L.; Wang, E.; Weaver, S.C. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: Infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology 2008, 372, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Kenney, J.L.; Adams, A.P.; Gorchakov, R.; Leal, G.; Weaver, S.C. Genetic and anatomic determinants of enzootic Venezuelan equine encephalitis virus infection of Culex (Melanoconion) taeniopus. PLoS Negl. Trop. Dis. 2012, 6, e1606. [Google Scholar] [CrossRef]
- Brackney, D.E.; Brown, I.K.; Nofchissey, R.A.; Fitzpatrick, K.A.; Ebel, G.D. Homogeneity of Powassan virus populations in naturally infected Ixodes scapularis. Virology 2010, 402, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Beeharry, Y.; Borderia, A.V.; Blanc, H.; Vignuzzi, M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. 2011, 108, 16038–16043. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies diversity determines pathogenesis through co-operative interactions in a viral population. Nature 2006, 439, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, J.K.; Kirkegaard, K. Increased fidelity reduces Poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005, 1, e11. [Google Scholar] [CrossRef] [PubMed]
- Rosen-Gagnon, K.; Stapleford, K.A.; Mongelli, V.; Blanc, H.; Failloux, A.-B.; Saleh, M.-C.; Vignuzzi, M. Alphavirus mutator variants present host-specific defects and attenuation in mammalian and insect models. PLoS Pathog. 2014, 10, e1003877. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.J.; Myles, K.M.; Raikhel, A.S. Small RNAs: A new frontier in mosquito biology. Trends Parasitol. 2013, 29, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Fragkoudis, R.; Attarzadeh-Yazdi, G.; Nash, A.A.; Fazakerley, J.K.; Kohl, A. Advances in dissecting mosquito innate immune responses to arbovirus infection. J. Gen. Virol. 2009, 90, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.D.; Sanchez-Vargas, I.; Franz, A.W.E.; Olson, K.E. Rendering mosquitoes resistant to Arboviruses through RNA interference. Microbe 2006, 1, 466–470. [Google Scholar]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. 2004, 101, 17240–17245. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Popli, S.; Hari, Y.; Malhotra, P.; Mukherjee, S.; Bhatnagar, R.K. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 2009, 23, 1845–1857. [Google Scholar] [CrossRef] [PubMed]
- Cirimotich, C.M.; Scott, J.C.; Phillips, A.T.; Geiss, B.J.; Olson, K.E. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Morazzani, E.M.; Adelman, Z.N. Origins of alphavirus-derived small RNA’s in mosquitoes. RNA Biol. 2009, 6, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. 2008, 105, 19938–19943. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Pesko, K.N.; Brown, I.K.; Deardorff, E.R.; Kawatachi, J.; Ebel, G.D. West Nile virus genetic diversity is maintained during transmission by Culex pipiens quinquefasciatus mosquitoes. PLoS One 2011, 6, e24466. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.E.; Morales, N.M.; Alto, B.W.; Remold, S.K. Role of evolved host breadth in the initial emergence of an RNA virus. Evolution 2010, 64, 3273–3286. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.T.; Ebel, G.D.; Lanciotti, R.S.; Brault, A.C.; Guzman, H.; Siirin, M.; Lambert, A.; Parsons, R.E.; Beasley, D.W.; Novak, R.J.; et al. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology 2005, 342, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Ebel, G.D.; Carricaburu, J.; Young, D.; Bernard, K.A.; Kramer, L.D. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am. J. Trop. Med. Hyg. 2004, 71, 493–500. [Google Scholar] [PubMed]
- Parameswaran, P.; Charlebois, P.; Tellez, Y.; Nunez, A.; Ryan, E.M.; Malboeuf, C.M.; Levin, J.Z.; Lennon, N.J.; Balmaseda, A.; Harris, E.; et al. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J. Virol. 2012, 86, 8546–8558. [Google Scholar] [CrossRef] [PubMed]
- Ciota, A.T.; Ehrbar, D.J.; Matacchiero, A.C.; van Slyke, G.A.; Kramer, L.D. The evolution of virulence of West Nile virus in a mosquito vector: Implications for arbovirus adaptation and evolution. BMC Evol. Biol. 2013, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Chen, R.; Sherman, M.B.; Weaver, S.C. Chikungunya virus: Evolution and genetic determinants of emergence. Curr. Opin. Virol. 2011, 1, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Hall-Mendelin, S.; Allcock, R.; Kresoje, N.; van den Hurk, A.F.; Warrilow, D. Detection of arboviruses and other micro-organisms in experimentally infected mosquitoes using massively parallel sequencing. PLoS One 2013, 8, e58026. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Ferro, C.; Barrera, R.; Boshell, J.; Navarro, J.C. Venezuelan equine encephalitis. Ann. Rev. Entomol. 2004, 49, 141–174. [Google Scholar] [CrossRef]
- May, F.J.; Davis, C.T.; Tesh, R.B.; Barrett, A.D. Phylogeography of West Nile virus: From the cradle of evolution in Africa to Eurasia, Australia and the Americas. J. Virol. 2011, 85, 2964–2974. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.T.; Beasley, D.W.; Guzman, H.; Raj, R.; D’Anton, M.; Novak, R.J.; Unnasch, T.R.; Tesh, R.B.; Barrett, A.D. Genetic variation among temporally and geographically distinct West Nile virus isolates, United States, 2001, 2002. Emerg. Infect. Dis. 2003, 9, 1423–1429. [Google Scholar] [CrossRef]
- Mann, B.R.; McMullen, A.R.; Swetnam, D.M.; Barrett, A.D. Molecular epidemiology and evolution of West Nile virus in North America. Int. J. Environ. Res. Public Health 2013, 10, 5111–5129. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.R.; McMullen, A.R.; Swetnam, D.M.; Salvato, V.; Reyna, M.; Guzman, H.; Bueno, R., Jr.; Dennett, J.A.; Tesh, R.B.; Barrett, A.D. Continued evolution of West Nile virus, Houston, Texas, USA, 2002–2012. Emerg. Infect. Dis. 2013, 19, 1418–1427. [Google Scholar] [CrossRef] [PubMed]
- McMullen, A.R.; Albayrak, H.; May, F.J.; Davis, C.T.; Beasley, D.W.; Barrett, A.D. Molecular evolution of lineage 2 West Nile virus. J. Gen. Virol. 2013, 94, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Jerzak, G.; Bernard, K.; Kramer, L.; Shi, P.-Y.; Ebel, G. The West Nile virus mutant spectrum is host-dependent and a determinant of mortality in mice. Virology 2007, 360, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, E.R.; FItzpartrick, K.A.; Jerzak, G.; Shi, P.Y.; Kramer, L.D.; Ebel, G.D. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog. 2011, 7, e1002335. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Vasilakis, N.; Brault, A.C.; Powers, A.M.; Tripet, F.; Weaver, S.C. Arbovirus evolution in vivo is constrained by host alternation. Proc. Natl. Acad. Sci. 2008, 105, 6970–6975. [Google Scholar] [CrossRef] [PubMed]
- Ciota, A.; Ehrbar, D.J.; Van Slyke, G.A.; Payne, A.F.; Willsey, G.G.; Viscio, R.E.; Kramer, L.D. Quantification of intrahost bottlenecks of West Nile virus in Culex pipiens mosquitoes using an artificial mutant swarm. Infect. Genet. Evol. 2012, 12, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, E.R.; Forrester, N.L.; Travassos-da-Rosa, A.P.; Estrada-Franco, J.G.; Navarro-Lopez, R.; Tesh, R.B.; Weaver, S.C. Experimental infection of potential reservoir hosts with Venezuelan equine encephalitis virus, Mexico. Emerg. Infect. Dis. 2009, 15, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Failloux, A.-B.; Weaver, S.C. Chikungunya Virus-vector Interactions. Viruses 2014, 6. in press. [Google Scholar]
- Coffey, L.L.; Vignuzzi, M. Host alternation of chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J. Virol. 2011, 85, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Stapleford, K.A.; Coffey, L.L.; Lay, S.; Borderia, A.V.; Duong, V.; Isakov, O.; Rozen-Gagnon, K.; Arias-Goeta, C.; Blanc, H.; Beaucourt, S.; et al. Emergence and transmission of arbovirus evolutionary intermediates with epidemic potential. Cell Host Microbe 2014, 15, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic control. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Vazeille, M.; Moutailler, S.; Coudrier, D.; Rousseaux, C.; Khun, H.; Huerre, M.; Thiria, J.; Dehecq, J.S.; Fontenille, D.; Schuffenecker, I.; et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One 2007, 2, e1168. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forrester, N.L.; Coffey, L.L.; Weaver, S.C. Arboviral Bottlenecks and Challenges to Maintaining Diversity and Fitness during Mosquito Transmission. Viruses 2014, 6, 3991-4004. https://doi.org/10.3390/v6103991
Forrester NL, Coffey LL, Weaver SC. Arboviral Bottlenecks and Challenges to Maintaining Diversity and Fitness during Mosquito Transmission. Viruses. 2014; 6(10):3991-4004. https://doi.org/10.3390/v6103991
Chicago/Turabian StyleForrester, Naomi L., Lark L. Coffey, and Scott C. Weaver. 2014. "Arboviral Bottlenecks and Challenges to Maintaining Diversity and Fitness during Mosquito Transmission" Viruses 6, no. 10: 3991-4004. https://doi.org/10.3390/v6103991



