Generation and Characterization of a Novel Recombinant Antibody against LMP1-TES1 of Epstein-Barr Virus Isolated by Phage Display
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Selection of Specific LMP1 Binding Phage and Nucleic Acid Analysis of htesFab Clones
| Fab library | 1st round | 2nd round | 3rd round |
|---|---|---|---|
| Phage input (cfu) | 2.0 × 1012 | 2.0 × 1010 | 1.0 × 1010 |
| Phage output (cfu) | 2.0 × 105 | 2.5 × 105 | 1.0 × 105 |
| Output/input | 1.0 × 10−7 | 1.25 × 10−5 | 1.6 × 10−5 |
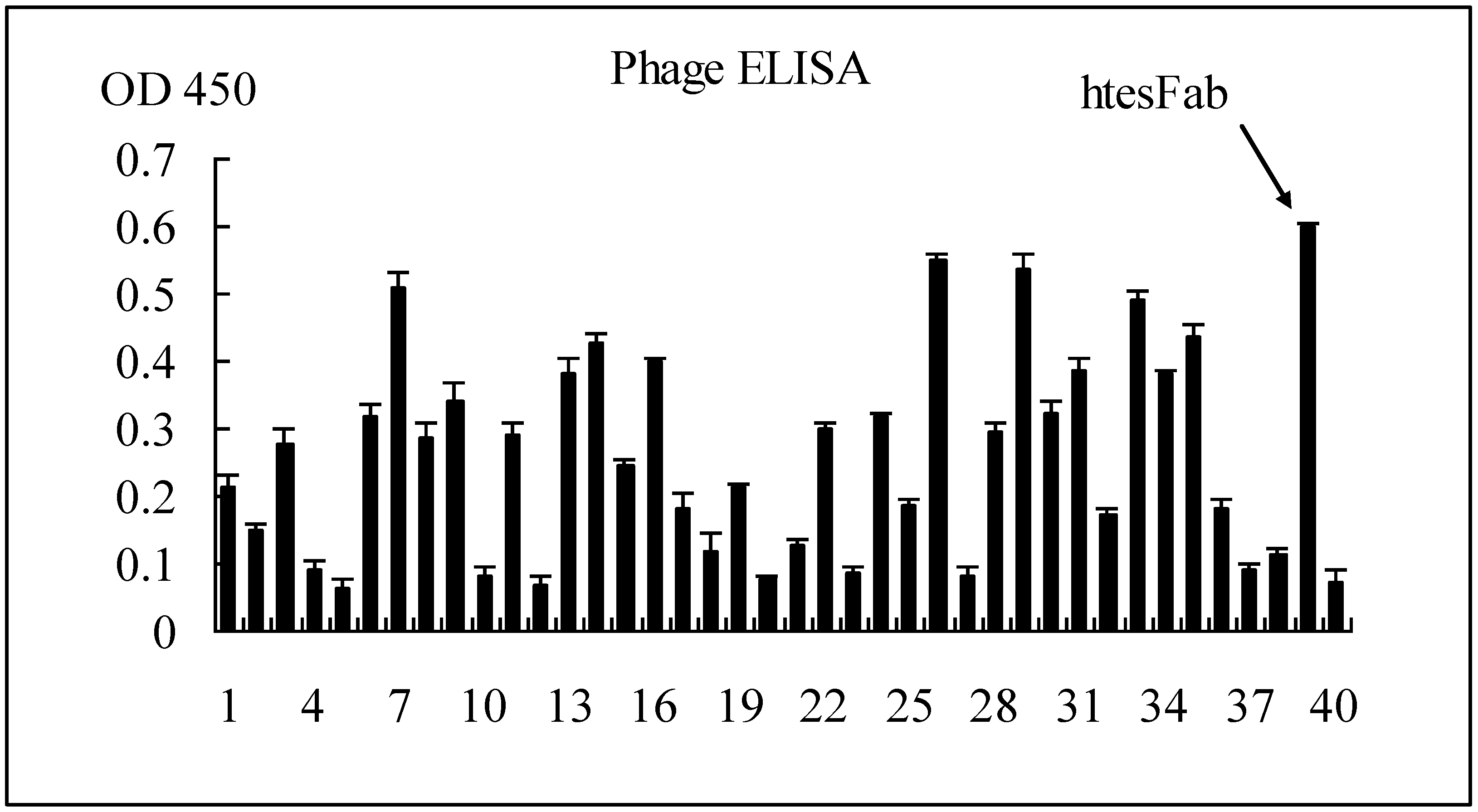
 |
2.1.2. Expression and Purification of htesFab

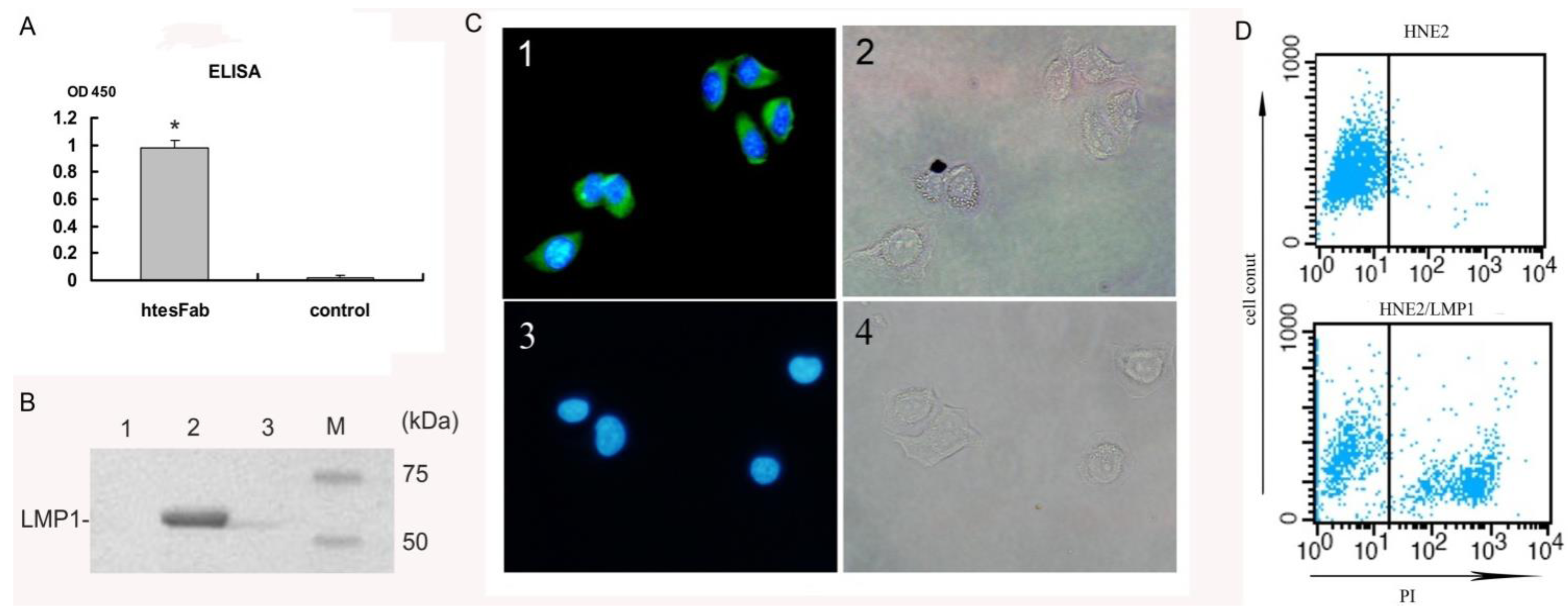
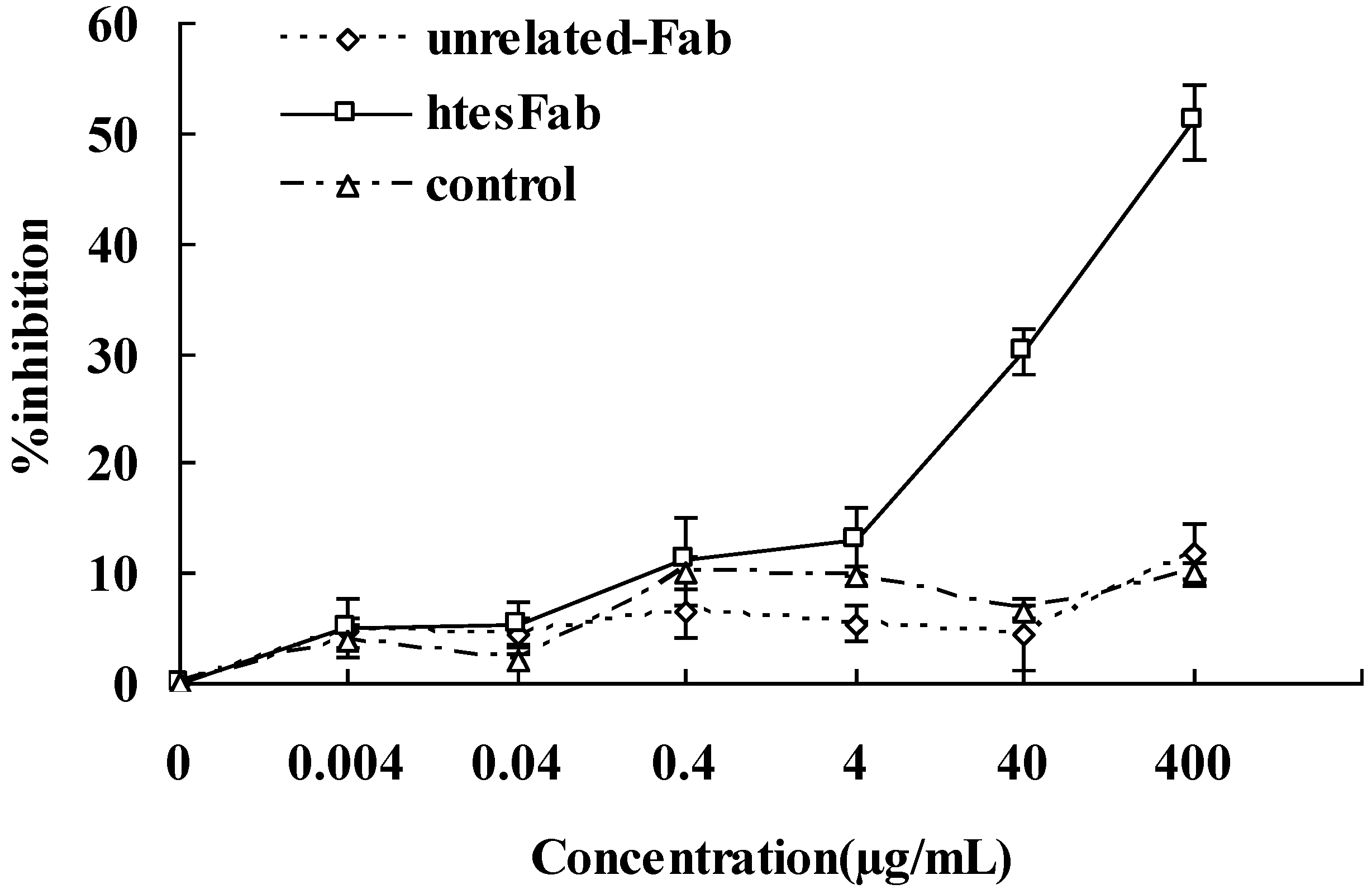
2.2. Discussion
3. Experimental
3.1. Phage Library, Helper Phage and Bacterial Strains
3.2. Cell Lines and Peptides
3.3. Bio-Panning
3.4. ELISA Screening of LMP1-Binding-Positive Phage Clones
3.5. Nucleic Acid Analysis of htesFab Clones
3.6. Expression and Purification of a Soluble Fab Fragment
3.7. Immunoprecipitation
3.8. Immunofluorescence Assay
3.9. Fluorescence-Activated Cell Sorter Analysis (FACS)
3.10. MTT Assay
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Masmoudi, A.; Toumi, N.; Khanfir, A.; Kallel-Slimi, L.; Daoud, J.; Karray, H.; Frikha, M. Epstein-Barr virus-targeted immunotherapy for nasopharyngeal carcinoma. Cancer Treat Rev. 2007, 33, 499–505. [Google Scholar] [CrossRef]
- Frappier, L. Contributions of Epstein-Barr nuclear antigen 1 (EBNA1) to cell immortalization and survival. Viruses 2012, 14, 1537–1547. [Google Scholar] [CrossRef]
- Hu, L.F.; Zhen, Q.F.; Zhang, Y.W.; Luo, Y.; Zheng, X.; Winberg, G.; Ernberg, I.; Klein, G. Differences in the growth pattern and clinical course of EBV-LMP1 expressing and nonexpressing nasopharyngeal carcinoma. Eur. J. Cancer 1995, 31, 658–660. [Google Scholar]
- Zhao, Y.; Wang, Y.; Zeng, S.; Hu, X. LMP1 expression is positively associated with metastasis of nasopharyngeal carcinoma: Evidence from a meta-analysis. J. Clin. Pathol. 2012, 65, 41–45. [Google Scholar]
- Li, H.P.; Chang, Y.S. Epstein-Barr virus latent membrane protein 1: Structure and functions. J. Biomed. Sci. 2003, 10, 490–504. [Google Scholar] [CrossRef]
- Moorthy, R.K.; Thorley-Lawson, D.A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required fortransformation of rat-1 fibroblasts. J. Virol. 1993, 67, 1638–1646. [Google Scholar]
- Coffin, W.F., 3rd; Erickson, K.D.; Hoedt-Miller, M.; Martin, J.M. The cytoplasmic amino-terminus of the Latent Membrane Protein-1 of Epstein-Barr Virus: Relationship between transmembrane orientation and effector functions of the carboxy-terminus and transmembrane domain. Oncogene 2001, 20, 5313–5330. [Google Scholar]
- Eliopoulos, A.G.; Young, L.S. LMP1 structure and signal transduction. Semin. Cancer Biol. 2001, 11, 435–444. [Google Scholar] [CrossRef]
- Kaye, K.M.; Izumi, K.M.; Mosialos, G.; Kieff, E. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J. Virol. 1995, 69, 675–683. [Google Scholar]
- Mosialos, G.; Birkenbach, M.; Yalamanchili, R.; van Arsdale, T.; Ware, C.; Kieff, E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 1995, 80, 389–399. [Google Scholar] [CrossRef]
- Brodeur, S.R.; Cheng, G.; Baltimore, D.; Thorley-Lawson, D.A. Localization of the major NF-kappaB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J. Biol. Chem. 1997, 272, 19777–19784. [Google Scholar]
- Devergne, O.; McFarland, E.C.; Mosialos, G.; Izumi, K.M.; Ware, C.F.; Kieff, E. Role of the TRAF binding site and NF-kB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 1998, 72, 7900–7908. [Google Scholar]
- Sandberg, M.; Hammerschmidt, W.; Sugden, B. Characterization of LMP-1’s association with TRAF1, TRAF2, and TRAF3. J. Virol. 1997, 71, 4649–4656. [Google Scholar]
- Devergne, O.; Hatzivassiliou, E.; Izumi, K.M.; Kaye, K.M.; Kleijnen, M.F.; Kieff, E.; Mosialos, G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: Role in NF-kB activation. Mol. Cell. Biol. 1996, 16, 7098–7108. [Google Scholar]
- Soni, V.; Cahir-McFarland, E.; Kieff, E. LMP1 TRAFficking activates growth and survival pathways. Adv. Exp. Med. Biol. 2007, 597, 173–187. [Google Scholar]
- Cheng, H.M.; Foong, Y.T.; Sam, C.K.; Prasad, U.; Dillner, J. Epstein-Barr virus nuclear antigen 1 linear epitopes that are reactive with immunoglobulin A (IgA) or IgG in sera from nasopharyngeal carcinoma patients or from healthy donors. J. Clin. Microbiol. 1991, 29, 2180–2186. [Google Scholar]
- Yip, T.T.; Ngan, R.K.; Lau, W.H.; Poon, Y.F.; Joab, I.; Cochet, C.; Cheng, A.K. A possible prognostic role of immunoglobulin-G antibody against recombinant Epstein-Barr virus BZLF-1 transactivator protein ZEBRA in patients with nasopharyngeal carcinoma. Cancer 1994, 74, 2414–2424. [Google Scholar] [CrossRef]
- Tang, J.W.; Rohwader, E.; Chu, I.M.; Tsang, R.K.; Steinhagen, K.; Yeung, A.C.; To, K.F.; Chan, P.K. Evaluation of Epstein-Barr virus antigen-based immunoassays for serological diagnosis of nasopharyngeal carcinoma. J. Clin. Virol. 2007, 40, 248–284. [Google Scholar] [CrossRef]
- Paramita, D.K.; Fachiroh, J.; Haryana, S.M.; Middeldorp, J.M. Evaluation of commercial EBV Recombline assay for diagnosis of nasopharyngeal carcinoma. J. Clin. Virol. 2008, 42, 343–352. [Google Scholar] [CrossRef]
- Cho, W.C. Nasopharyngeal carcinoma: Molecular biomarker discovery and progress. Mol. Cancer 2007, 6, 1–9. [Google Scholar] [CrossRef]
- Ai, P.; Wang, T.; Zhang, H.; Wang, Y.; Song, C.; Zhang, L.; Li, Z.; Hu, H. Determination of antibodies directed at EBV proteins expressed in both latent and lytic cycles in nasopharyngeal carcinoma. Oral Oncol. 2013, 4, 326–331. [Google Scholar]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Azarnia, N.; Shin, D.M.; Cohen, R.B.; Jones, C.U.; Sur, R.; Raben, D.; Jassem, J.; et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006, 354, 567–578. [Google Scholar] [CrossRef]
- Spratt, D.E.; Lee, N. Current and emerging treatment options for nasopharyngeal carcinoma. Oncol. Targets Ther. 2012, 5, 297–308. [Google Scholar]
- Fang, C.Y.; Chang, Y.S.; Chow, K.P.; Yu, J.S.; Chang, H.Y. Construction and characterization of monoclonal antibodies specific to Epstein-Barr virus latent membrane protein 1. J. Immunol. Methods 2004, 287, 21–30. [Google Scholar] [CrossRef]
- Gennari, F.; Mehta, S.; Wang, Y.; St Clair Tallarico, A.; Palu, G.; Marasco, W.A. Direct phage to intrabody screening (DPIS): Demonstration by isolation of cytosolic intrabodies against the TES1 site of Epstein Barr virus latent membrane protein 1 (LMP1) that block NF-kappaB transactivation. J. Mol. Biol. 2004, 335, 193–207. [Google Scholar] [CrossRef]
- Piché, A.; Kasono, K.; Johanning, F.; Curiel, T.J.; Curiel, D.T. Phenotypic knock-out of the latent membrane protein 1 of Epstein-Barr virus by an intracellular single-chain antibody. Gene Ther. 1998, 5, 1171–1179. [Google Scholar]
- Biocca, S.; Pierandrei-Amaldi, P.; Cattaneo, A. Intracellular expression of anti-p21ras single chain Fv fragments inhibits meiotic maturation of Xenopus oocytes. Biochem. Biophys. Res. Commun. 1993, 197, 422–427. [Google Scholar] [CrossRef]
- Tavladoraki, P.; Benvenuto, E.; Trinca, S.; De Martinis, D.; Cattaneo, A.; Galeffi, P. Transgenic plants expressing a functional singlechain Fv antibody are specifically protected from virus attack. Nature 1993, 366, 469–472. [Google Scholar]
- Rondon, I.J.; Marasco, W.A. Intracellular antibodies (intrabodies) for gene therapy of infectious diseases. Annu. Rev. Microbiol. 1997, 51, 257–283. [Google Scholar] [CrossRef]
- Lin, H.; Mao, Y.; Zhang, D.W.; Li, H.; Qiu, J.R.; Zhu, J.; Chen, R.J. Selection and Characterization of human Anti-MAGE-A1 scFv and immunotoxin. Anticancer Agents Med. Chem. 2013, in press. [Google Scholar]
- Cattaneo, A.; Biocca, S. The selection of intracellular antibodies. Trends Biotechnol. 1999, 17, 115–121. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhao, P.; Zhu, J.; Grabinski, T.; Feng, Z.; Guan, X.; Skinner, R.S.; Gross, M.D.; Hay, R.V.; Tachibana, H.; et al. Construction of human naive Fab library and characterization of anti-met Fab fragment generated from the library. Mol. Biotechnol. 2005, 31, 41–54. [Google Scholar]
- IMGT/LIGM Database. Available online: http://www.imgt.org/IMGTlect/ (accessed on 26 March 2013).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, D.; Mao, Y.; Cao, Q.; Xiong, L.; Wen, J.; Chen, R.; Zhu, J. Generation and Characterization of a Novel Recombinant Antibody against LMP1-TES1 of Epstein-Barr Virus Isolated by Phage Display. Viruses 2013, 5, 1131-1142. https://doi.org/10.3390/v5041131
Zhang D, Mao Y, Cao Q, Xiong L, Wen J, Chen R, Zhu J. Generation and Characterization of a Novel Recombinant Antibody against LMP1-TES1 of Epstein-Barr Virus Isolated by Phage Display. Viruses. 2013; 5(4):1131-1142. https://doi.org/10.3390/v5041131
Chicago/Turabian StyleZhang, Dawei, Yuan Mao, Qing Cao, Lin Xiong, Juan Wen, Renjie Chen, and Jin Zhu. 2013. "Generation and Characterization of a Novel Recombinant Antibody against LMP1-TES1 of Epstein-Barr Virus Isolated by Phage Display" Viruses 5, no. 4: 1131-1142. https://doi.org/10.3390/v5041131




