Breaking In: Human Metapneumovirus Fusion and Entry
Abstract
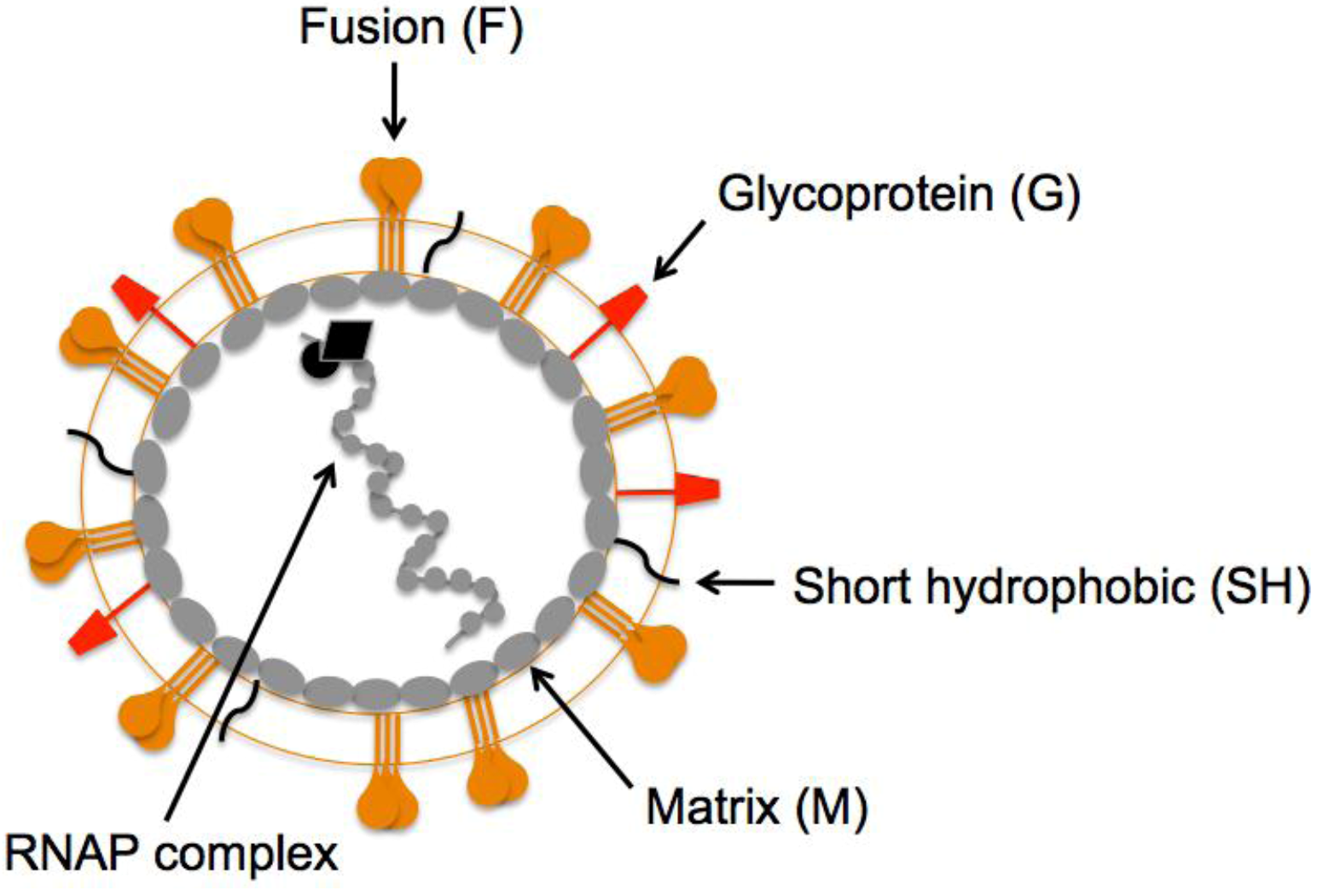
:1. Introduction
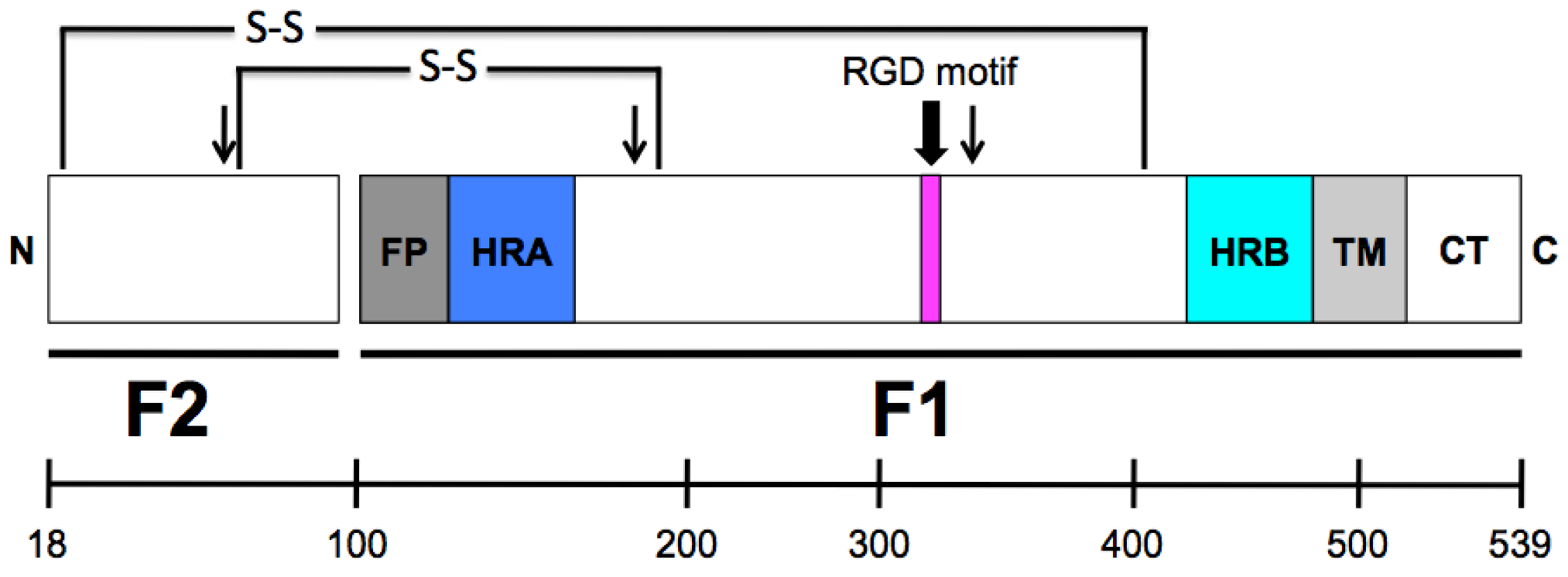
2. HMPV F: A Dual Function Fusion Protein
3. HMPV F: The Key to Intrusion
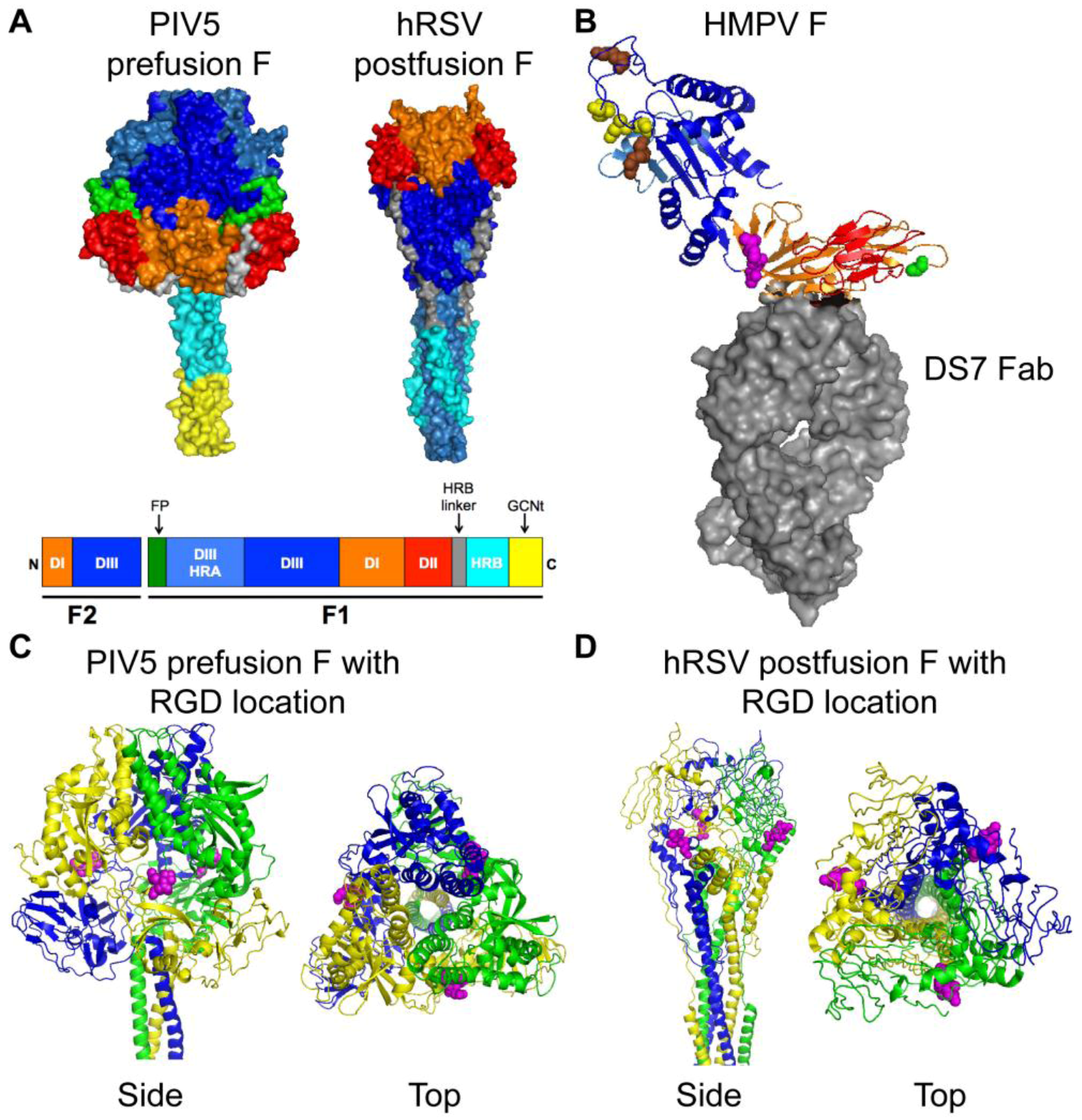
4. HMPV Binding: Identifying the Door
5. HMPV Fusion: Entering the Premises
6. HMPV Entry: Unlocking the Mystery
7. Conclusions
Acknowledgments
Conflict of Interest
References
- van den Hoogen, B.G.; de Jong, J.C.; Groen, J.; Kuiken, T.; de Groot, R.; Fouchier, R.A.; Osterhaus, A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001, 7, 719–724. [Google Scholar] [CrossRef]
- Schildgen, V.; van den Hoogen, B.; Fouchier, R.; Tripp, R.A.; Alvarez, R.; Manoha, C.; Williams, J.; Schildgen, O. Human metapneumovirus: Lessons learned over the first decade. Clin. Microbiol. Rev. 2011, 24, 734–754. [Google Scholar] [CrossRef]
- Cook, J.K. Avian pneumovirus infections of turkeys and chickens. Vet. J. 2000, 160, 118–125. [Google Scholar]
- van den Hoogen, B.G.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Analysis of the genomic sequence of a human metapneumovirus. Virology 2002, 295, 119–132. [Google Scholar] [CrossRef]
- Williams, J.V.; Harris, P.A.; Tollefson, S.J.; Halburnt-Rush, L.L.; Pingsterhaus, J.M.; Edwards, K.M.; Wright, P.F.; Crowe, J.E., Jr. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 2004, 350, 443–450. [Google Scholar]
- de Graaf, M.; Osterhaus, A.D.; Fouchier, R.A.; Holmes, E.C. Evolutionary dynamics of human and avian metapneumoviruses. J. Gen. Virol. 2008, 89, 2933–2942. [Google Scholar]
- Yang, C.F.; Wang, C.K.; Tollefson, S.J.; Piyaratna, R.; Lintao, L.D.; Chu, M.; Liem, A.; Mark, M.; Spaete, R.R.; Crowe, J.E., Jr.; et al. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol. J. 2009, 6, 138. [Google Scholar] [CrossRef]
- Williams, J.V.; Wang, C.K.; Yang, C.F.; Tollefson, S.J.; House, F.S.; Heck, J.M.; Chu, M.; Brown, J.B.; Lintao, L.D.; Quinto, J.D.; et al. The role of human metapneumovirus in upper respiratory tract infections in children: A 20-year experience. J. Infect. Dis. 2006, 193, 387–395. [Google Scholar] [CrossRef]
- van den Hoogen, B.G.; van Doornum, G.J.; Fockens, J.C.; Cornelissen, J.J.; Beyer, W.E.; de Groot, R.; Osterhaus, A.D.; Fouchier, R.A. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 2003, 188, 1571–1577. [Google Scholar] [CrossRef]
- Peiris, J.S.; Tang, W.H.; Chan, K.H.; Khong, P.L.; Guan, Y.; Lau, Y.L.; Chiu, S.S. Children with respiratory disease associated with metapneumovirus in hong kong. Emerg. Infect. Dis. 2003, 9, 628–633. [Google Scholar] [CrossRef]
- Mullins, J.A.; Erdman, D.D.; Weinberg, G.A.; Edwards, K.; Hall, C.B.; Walker, F.J.; Iwane, M.; Anderson, L.J. Human metapneumovirus infection among children hospitalized with acute respiratory illness. Emerg. Infect. Dis. 2004, 10, 700–705. [Google Scholar]
- McAdam, A.J.; Hasenbein, M.E.; Feldman, H.A.; Cole, S.E.; Offermann, J.T.; Riley, A.M.; Lieu, T.A. Human metapneumovirus in children tested at a tertiary-care hospital. J. Infect. Dis. 2004, 190, 20–26. [Google Scholar] [CrossRef]
- Mackay, I.M.; Bialasiewicz, S.; Jacob, K.C.; McQueen, E.; Arden, K.E.; Nissen, M.D.; Sloots, T.P. Genetic diversity of human metapneumovirus over 4 consecutive years in australia. J. Infect. Dis. 2006, 193, 1630–1633. [Google Scholar] [CrossRef]
- Foulongne, V.; Guyon, G.; Rodiere, M.; Segondy, M. Human metapneumovirus infection in young children hospitalized with respiratory tract disease. Pediatr. Infect. Dis. J. 2006, 25, 354–359. [Google Scholar] [CrossRef]
- Esper, F.; Martinello, R.A.; Boucher, D.; Weibel, C.; Ferguson, D.; Landry, M.L.; Kahn, J.S. A 1-year experience with human metapneumovirus in children aged <5 years. J. Infect. Dis. 2004, 189, 1388–1396. [Google Scholar] [CrossRef]
- Ebihara, T.; Endo, R.; Kikuta, H.; Ishiguro, N.; Ishiko, H.; Hara, M.; Takahashi, Y.; Kobayashi, K. Human metapneumovirus infection in japanese children. J. Clin. Microbiol. 2004, 42, 126–132. [Google Scholar]
- Dollner, H.; Risnes, K.; Radtke, A.; Nordbo, S.A. Outbreak of human metapneumovirus infection in norwegian children. Pediatr. Infect. Dis. J. 2004, 23, 436–440. [Google Scholar] [CrossRef]
- Boivin, G.; De Serres, G.; Cote, S.; Gilca, R.; Abed, Y.; Rochette, L.; Bergeron, M.G.; Dery, P. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 2003, 9, 634–640. [Google Scholar] [CrossRef]
- Williams, J.V.; Martino, R.; Rabella, N.; Otegui, M.; Parody, R.; Heck, J.M.; Crowe, J.E., Jr. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J. Infect. Dis. 2005, 192, 1061–1065. [Google Scholar] [CrossRef]
- Williams, J.V.; Crowe, J.E., Jr.; Enriquez, R.; Minton, P.; Peebles, R.S., Jr.; Hamilton, R.G.; Higgins, S.; Griffin, M.; Hartert, T.V. Human metapneumovirus infection plays an etiologic role in acute asthma exacerbations requiring hospitalization in adults. J. Infect. Dis. 2005, 192, 1149–1153. [Google Scholar] [CrossRef]
- Vicente, D.; Montes, M.; Cilla, G.; Perez-Trallero, E. Human metapneumovirus and chronic obstructive pulmonary disease. Emerg. Infect. Dis. 2004, 10, 1338–1339. [Google Scholar] [CrossRef]
- Pelletier, G.; Dery, P.; Abed, Y.; Boivin, G. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 2002, 8, 976–978. [Google Scholar] [CrossRef]
- Madhi, S.A.; Ludewick, H.; Abed, Y.; Klugman, K.P.; Boivin, G. Human metapneumovirus-associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (hiv-1)-infected and hiv-1-uninfected african infants. Clin. Infect. Dis. 2003, 37, 1705–1710. [Google Scholar] [CrossRef]
- Larcher, C.; Geltner, C.; Fischer, H.; Nachbaur, D.; Muller, L.C.; Huemer, H.P. Human metapneumovirus infection in lung transplant recipients: Clinical presentation and epidemiology. J. Heart. Lung. Transplant. 2005, 24, 1891–1901. [Google Scholar] [CrossRef]
- Englund, J.A.; Boeckh, M.; Kuypers, J.; Nichols, W.G.; Hackman, R.C.; Morrow, R.A.; Fredricks, D.N.; Corey, L. Brief communication: Fatal human metapneumovirus infection in stem-cell transplant recipients. Ann. Intern. Med. 2006, 144, 344–349. [Google Scholar]
- Brodzinski, H.; Ruddy, R.M. Review of new and newly discovered respiratory tract viruses in children. Pediatr. Emerg. Care 2009, 25, 352–360. [Google Scholar] [CrossRef]
- Loughlin, G.M.; Moscona, A. The cell biology of acute childhood respiratory disease: Therapeutic implications. Pediatr. Clin. North. Am. 2006, 53, 929–959. [Google Scholar] [CrossRef]
- Kuiken, T.; van den Hoogen, B.G.; van Riel, D.A.; Laman, J.D.; van Amerongen, G.; Sprong, L.; Fouchier, R.A.; Osterhaus, A.D. Experimental human metapneumovirus infection of cynomolgus macaques (macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. Am. J. Pathol. 2004, 164, 1893–1900. [Google Scholar] [CrossRef]
- Williams, J.V.; Tollefson, S.J.; Johnson, J.E.; Crowe, J.E., Jr. The cotton rat (sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J. Virol. 2005, 79, 10944–10951. [Google Scholar] [CrossRef]
- Hamelin, M.E.; Prince, G.A.; Gomez, A.M.; Kinkead, R.; Boivin, G. Human metapneumovirus infection induces long-term pulmonary inflammation associated with airway obstruction and hyperresponsiveness in mice. J. Infect. Dis. 2006, 193, 1634–1642. [Google Scholar] [CrossRef]
- Roberts, S.R.; Compans, R.W.; Wertz, G.W. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 1995, 69, 2667–2673. [Google Scholar]
- Shaikh, F.Y.; Cox, R.G.; Lifland, A.W.; Hotard, A.L.; Williams, J.V.; Moore, M.L.; Santangelo, P.J.; Crowe, J.E., Jr. A critical phenylalanine residue in the respiratory syncytial virus fusion protein cytoplasmic tail mediates assembly of internal viral proteins into viral filaments and particles. MBio 2012, 3. [Google Scholar] [CrossRef]
- Sabo, Y.; Ehrlich, M.; Bacharach, E. The conserved yagl motif in human metapneumovirus is required for higher-order cellular assemblies of the matrix protein and for virion production. J. Virol. 2011, 85, 6594–6609. [Google Scholar]
- Lamb, R.A. Paramyxovirus fusion: A hypothesis for changes. Virology 1993, 197, 1–11. [Google Scholar] [CrossRef]
- Karron, R.A.; Buonagurio, D.A.; Georgiu, A.F.; Whitehead, S.S.; Adamus, J.E.; Clements-Mann, M.L.; Harris, D.O.; Randolph, V.B.; Udem, S.A.; Murphy, B.R.; et al. Respiratory syncytial virus (rsv) sh and g proteins are not essential for viral replication in vitro: Clinical evaluation and molecular characterization of a cold-passaged, attenuated rsv subgroup b mutant. P. Natl. Acad. Sci. USA 1997, 94, 13961–13966. [Google Scholar]
- Feldman, S.A.; Audet, S.; Beeler, J.A. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 2000, 74, 6442–6447. [Google Scholar]
- Kahn, J.S.; Schnell, M.J.; Buonocore, L.; Rose, J.K. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (rsv) glycoproteins: Rsv fusion protein can mediate infection and cell fusion. Virology 1999, 254, 81–91. [Google Scholar] [CrossRef]
- Schlender, J.; Zimmer, G.; Herrler, G.; Conzelmann, K.K. Respiratory syncytial virus (rsv) fusion protein subunit f2, not attachment protein g, determines the specificity of rsv infection. J. Virol. 2003, 77, 4609–4616. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Barretto, N.; Peeples, M.E. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 2001, 75, 6825–6834. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Collins, P.L.; Peeples, M.E. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 2002, 294, 296–304. [Google Scholar] [CrossRef]
- Widjojoatmodjo, M.N.; Boes, J.; van Bers, M.; van Remmerden, Y.; Roholl, P.J.; Luytjes, W. A highly attenuated recombinant human respiratory syncytial virus lacking the g protein induces long-lasting protection in cotton rats. Virol. J. 2010, 7, 114. [Google Scholar] [CrossRef]
- Teng, M.N.; Whitehead, S.S.; Collins, P.L. Contribution of the respiratory syncytial virus g glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 2001, 289, 283–296. [Google Scholar] [CrossRef]
- Biacchesi, S.; Pham, Q.N.; Skiadopoulos, M.H.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Infection of nonhuman primates with recombinant human metapneumovirus lacking the sh, g, or m2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 2005, 79, 12608–12613. [Google Scholar] [CrossRef]
- Biacchesi, S.; Skiadopoulos, M.H.; Yang, L.; Lamirande, E.W.; Tran, K.C.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Recombinant human metapneumovirus lacking the small hydrophobic sh and/or attachment g glycoprotein: Deletion of g yields a promising vaccine candidate. J. Virol. 2004, 78, 12877–12887. [Google Scholar] [CrossRef]
- Hallak, L.K.; Collins, P.L.; Knudson, W.; Peeples, M.E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 2000, 271, 264–275. [Google Scholar] [CrossRef]
- Krusat, T.; Streckert, H.J. Heparin-dependent attachment of respiratory syncytial virus (rsv) to host cells. Arch. Virol. 1997, 142, 1247–1254. [Google Scholar] [CrossRef]
- Thammawat, S.; Sadlon, T.A.; Hallsworth, P.G.; Gordon, D.L. Role of cellular glycosaminoglycans and charged regions of viral g protein in human metapneumovirus infection. J. Virol. 2008, 82, 11767–11774. [Google Scholar] [CrossRef]
- Lamb, R.A.; Jardetzky, T.S. Structural basis of viral invasion: Lessons from paramyxovirus f. Curr. Opin. Struct. Biol. 2007, 17, 427–436. [Google Scholar] [CrossRef]
- Russell, C.J.; Jardetzky, T.S.; Lamb, R.A. Membrane fusion machines of paramyxoviruses: Capture of intermediates of fusion. EMBO J 2001, 20, 4024–4034. [Google Scholar] [CrossRef]
- Miller, S.A.; Tollefson, S.; Crowe, J.E., Jr.; Williams, J.V.; Wright, D.W. Examination of a fusogenic hexameric core from human metapneumovirus and identification of a potent synthetic peptide inhibitor from the heptad repeat 1 region. J. Virol. 2007, 81, 141–149. [Google Scholar] [CrossRef]
- Deffrasnes, C.; Hamelin, M.E.; Prince, G.A.; Boivin, G. Identification and evaluation of a highly effective fusion inhibitor for human metapneumovirus. Antimicrob. Agents Chemother. 2008, 52, 279–287. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Yin, H.S.; Wen, X.; Paterson, R.G.; Lamb, R.A.; Jardetzky, T.S. Structure of the parainfluenza virus 5 f protein in its metastable, prefusion conformation. Nature 2006, 439, 38–44. [Google Scholar]
- Swanson, K.A.; Settembre, E.C.; Shaw, C.A.; Dey, A.K.; Rappuoli, R.; Mandl, C.W.; Dormitzer, P.R.; Carfi, A. Structural basis for immunization with postfusion respiratory syncytial virus fusion f glycoprotein (rsv f) to elicit high neutralizing antibody titers. P. Natl. Acad. Sci. USA 2011, 108, 9619–9624. [Google Scholar]
- Wen, X.; Krause, J.C.; Leser, G.P.; Cox, R.G.; Lamb, R.A.; Williams, J.V.; Crowe, J.E., Jr.; Jardetzky, T.S. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat. Struct. Mol. Biol. 2012, 19, 461–463. [Google Scholar] [CrossRef]
- van den Hoogen, B.G.; Herfst, S.; Sprong, L.; Cane, P.A.; Forleo-Neto, E.; de Swart, R.L.; Osterhaus, A.D.; Fouchier, R.A. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 2004, 10, 658–666. [Google Scholar] [CrossRef]
- Boivin, G.; Mackay, I.; Sloots, T.P.; Madhi, S.; Freymuth, F.; Wolf, D.; Shemer-Avni, Y.; Ludewick, H.; Gray, G.C.; LeBlanc, E. Global genetic diversity of human metapneumovirus fusion gene. Emerg. Infect. Dis. 2004, 10, 1154–1157. [Google Scholar]
- Schowalter, R.M.; Smith, S.E.; Dutch, R.E. Characterization of human metapneumovirus f protein-promoted membrane fusion: Critical roles for proteolytic processing and low ph. J. Virol. 2006, 80, 10931–10941. [Google Scholar] [CrossRef]
- Zhang, J.; Dou, Y.; Wu, J.; She, W.; Luo, L.; Zhao, Y.; Liu, P.; Zhao, X. Effects of n-linked glycosylation of the fusion protein on replication of human metapneumovirus in vitro and in mouse lungs. J. Gen. Virol. 2011, 92, 1666–1675. [Google Scholar] [CrossRef]
- Cseke, G.; Maginnis, M.S.; Cox, R.G.; Tollefson, S.J.; Podsiad, A.B.; Wright, D.W.; Dermody, T.S.; Williams, J.V. Integrin alphavbeta1 promotes infection by human metapneumovirus. P. Natl. Acad. Sci. USA 2009, 106, 1566–1571. [Google Scholar]
- Chang, A.; Masante, C.; Buchholz, U.J.; Dutch, R.E. Human metapneumovirus (hmpv) binding and infection are mediated by interactions between the hmpv fusion protein and heparan sulfate. J. Virol. 2012, 86, 3230–3243. [Google Scholar] [CrossRef]
- de Graaf, M.; Schrauwen, E.J.; Herfst, S.; van Amerongen, G.; Osterhaus, A.D.; Fouchier, R.A. Fusion protein is the main determinant of metapneumovirus host tropism. J. Gen. Virol. 2009, 90, 1408–1416. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Stewart, P.L.; Nemerow, G.R. Cell integrins: Commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007, 15, 500–507. [Google Scholar] [CrossRef]
- Cox, R.G.; Livesay, S.B.; Johnson, M.; Ohi, M.D.; Williams, J.V. The human metapneumovirus fusion protein mediates entry via an interaction with rgd-binding integrins. J. Virol. 2012, 86, 12148–12160. [Google Scholar] [CrossRef]
- Wennerberg, K.; Lohikangas, L.; Gullberg, D.; Pfaff, M.; Johansson, S.; Fassler, R. Beta 1 integrin-dependent and -independent polymerization of fibronectin. J. Cell. Biol. 1996, 132, 227–238. [Google Scholar] [CrossRef]
- Fassler, R.; Pfaff, M.; Murphy, J.; Noegel, A.A.; Johansson, S.; Timpl, R.; Albrecht, R. Lack of beta 1 integrin gene in embryonic stem cells affects morphology, adhesion, and migration but not integration into the inner cell mass of blastocysts. J. Cell. Biol. 1995, 128, 979–988. [Google Scholar] [CrossRef]
- Tollefson, S.J.; Cox, R.G.; Williams, J.V. Studies of culture conditions and environmental stability of human metapneumovirus. Virus Res. 2010, 151, 54–59. [Google Scholar] [CrossRef]
- Shirogane, Y.; Takeda, M.; Iwasaki, M.; Ishiguro, N.; Takeuchi, H.; Nakatsu, Y.; Tahara, M.; Kikuta, H.; Yanagi, Y. Efficient multiplication of human metapneumovirus in vero cells expressing the transmembrane serine protease tmprss2. J. Virol. 2008, 82, 8942–8946. [Google Scholar] [CrossRef]
- Murakami, M.; Towatari, T.; Ohuchi, M.; Shiota, M.; Akao, M.; Okumura, Y.; Parry, M.A.; Kido, H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza a viruses and sendai virus. Eur. J. Biochem. 2001, 268, 2847–2855. [Google Scholar]
- Lamb, R.A.; Parks, G.D. Paramyxoviridae: The Viruses and their Replication. In Fields virology, 5th; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1, pp. 1449–1646. [Google Scholar]
- Herfst, S.; Mas, V.; Ver, L.S.; Wierda, R.J.; Osterhaus, A.D.; Fouchier, R.A.; Melero, J.A. Low-ph-induced membrane fusion mediated by human metapneumovirus f protein is a rare, strain-dependent phenomenon. J. Virol. 2008, 82, 8891–8895. [Google Scholar]
- Schowalter, R.M.; Chang, A.; Robach, J.G.; Buchholz, U.J.; Dutch, R.E. Low-ph triggering of human metapneumovirus fusion: Essential residues and importance in entry. J. Virol. 2009, 83, 1511–1522. [Google Scholar] [CrossRef]
- Mas, V.; Herfst, S.; Osterhaus, A.D.; Fouchier, R.A.; Melero, J.A. Residues of the human metapneumovirus fusion (f) protein critical for its strain-related fusion phenotype: Implications for the virus replication cycle. J. Virol. 2011, 85, 12650–12661. [Google Scholar] [CrossRef]
- Wei, Y.; Feng, K.; Yao, X.; Cai, H.; Li, J.; Mirza, A.M.; Iorio, R.M. Localization of a region in the fusion protein of avian metapneumovirus that modulates cell-cell fusion. J. Virol. 2012, 86, 11800–11814. [Google Scholar] [CrossRef]
- Chang, A.; Hackett, B.; Winter, C.C.; Buchholz, U.J.; Dutch, R.E. Potential electrostatic interactions in multiple regions affect hmpv f-mediated membrane fusion. J. Virol. 2012, 86, 9843–9853. [Google Scholar] [CrossRef]
- Williams, J.V.; Chen, Z.; Cseke, G.; Wright, D.W.; Keefer, C.J.; Tollefson, S.J.; Hessell, A.; Podsiad, A.; Shepherd, B.E.; Sanna, P.P.; et al. A recombinant human monoclonal antibody to human metapneumovirus fusion protein that neutralizes virus in vitro and is effective therapeutically in vivo. J. Virol. 2007, 81, 8315–8324. [Google Scholar]
- Swanson, K.; Wen, X.; Leser, G.P.; Paterson, R.G.; Lamb, R.A.; Jardetzky, T.S. Structure of the newcastle disease virus f protein in the post-fusion conformation. Virology 2010, 402, 372–379. [Google Scholar] [CrossRef]
- Yin, H.S.; Paterson, R.G.; Wen, X.; Lamb, R.A.; Jardetzky, T.S. Structure of the uncleaved ectodomain of the paramyxovirus (hpiv3) fusion protein. P. Natl. Acad. Sci. USA 2005, 102, 9288–9293. [Google Scholar]
- Tayyari, F.; Marchant, D.; Moraes, T.J.; Duan, W.; Mastrangelo, P.; Hegele, R.G. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat. Med. 2011, 17, 1132–1135. [Google Scholar]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. Molecular mechanisms of hiv entry. Adv. Exp. Med. Biol. 2012, 726, 223–242. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cox, R.G.; Williams, J.V. Breaking In: Human Metapneumovirus Fusion and Entry. Viruses 2013, 5, 192-210. https://doi.org/10.3390/v5010192
Cox RG, Williams JV. Breaking In: Human Metapneumovirus Fusion and Entry. Viruses. 2013; 5(1):192-210. https://doi.org/10.3390/v5010192
Chicago/Turabian StyleCox, Reagan G., and John V. Williams. 2013. "Breaking In: Human Metapneumovirus Fusion and Entry" Viruses 5, no. 1: 192-210. https://doi.org/10.3390/v5010192



