Immunity to Fish Rhabdoviruses
Abstract
:1. Introduction
2. The Fish Rhabdoviruses
2.1. Taxonomy
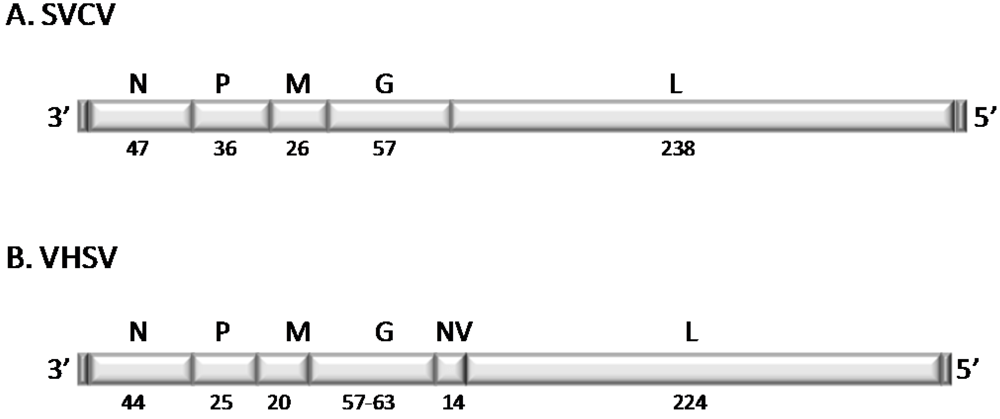
2.2. Structure
2.3. Infection Cycle
2.4. Disease Manifestations
2.5. Vaccination Strategies
3. Immune Response
3.2. Viral Recognition in Fish
3.3. Innate Immune Response to Fish Rhabdoviruses
3.4. Humoral Immune Response to Fish Rhabdoviruses
3.5. Cellular Immune Response to Fish Rhabdoviruses
3.6. Modulation of Immunity by Environmental Parameters
5. Future Directions
Acknowledgments
Conflict of Interest
References and Notes
- Kurath, G.; Winton, J. Fish Rhabdoviruses. In Encyclopedia of Virology, 3rd; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 221–227. [Google Scholar]
- Whitfield, A.E.; Calisher, C.H.; Stone, D.M.; Kurath, G.; Kuzmin, I.V.; Rodriguez, L.L.; Tordo, N.; Walker, P.J.; Dietzgen, R.G.; Tesh, R.B.; et al. Rhabdoviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; King, M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: Oxford, UK, 2011; pp. 686–714. [Google Scholar]
- Mork, C.; Hershberger, P.; Kocan, R.; Batts, W.; Winton, J. Isolation and characterization of a rhabdovirus from starry flounder (Platichthys stellatus) collected from the northern portion of Puget Sound, Washington, USA. J. Gen. Virol. 2004, 85, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Bootland, L.M.; Leong, J.C. Infectious haematopoietic necrosis virus. In Fish Diseases and Disorders; Woo, P.T.K., Bruno, D.W., Eds.; CAB International: Wallingford, UK, 1999; Volume 3, pp. 57–121. [Google Scholar]
- Ahne, W.; Bjorklund, H.V.; Essbauer, S.; Fijan, N.; Kurath, G.; Winton, J.R. Spring viremia of carp (SVC). Dis. Aquat. Organ. 2002, 52, 261–272. [Google Scholar]
- Al-Hussinee, L.; Lord, S.; Stevenson, R.M.; Casey, R.N.; Groocock, G.H.; Britt, K.L.; Kohler, K.H.; Wooster, G.A.; Getchell, R.G.; Bowser, P.R.; et al. Immunohistochemistry and pathology of multiple Great Lakes fish from mortality events associated with viral hemorrhagic septicemia virus type IVb. Dis. Aquat. Organ. 2011, 93, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.F. Genetic comparison of the rhabdoviruses from animals and plants. Curr. Top. Microbiol. Immunol. 2005, 292, 1–24. [Google Scholar]
- Wagner, R.R. Rhabdovirus biology and infection: An overview. In The Rhabdovirus; Wagner, R.R., Ed.; Plenum Press: New York, NY, USA, 1987; pp. 9–61. [Google Scholar]
- Hoffmann, B.; Schutze, H.; Mettenleiter, T.C. Determination of the complete genomic sequence and analysis of the gene products of the virus of Spring Viremia of Carp, a fish rhabdovirus. Virus Res. 2002, 84, 89–100. [Google Scholar]
- Kurath, G.; Leong, J.C. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. J. Virol. 1985, 53, 462–468. [Google Scholar]
- Schutze, H.; Mundt, E.; Mettenleiter, T.C. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 1999, 19, 59–65. [Google Scholar]
- Hilleman, M.R. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 14560–14566. [Google Scholar]
- LaPatra, S.E.; Lauda, K.A.; Jones, G.R.; Walker, S.C.; Shewmaker, B.S.; Morton, A.W. Characterization of IHNV isolates associated with neurotropism. Vet. Res. 1995, 26, 433–437. [Google Scholar]
- Ghittino, P. Viral hemorrhagic septicemia (VHS) in rainbow trout in Italy. Ann. N. Y. Acad. Sci. 1965, 126, 468–478. [Google Scholar]
- Vestergård Jørgensen, P.E. Egtved virus: Temperature-dependent immune response of trout to infection with low-virulence virus. J. Fish Dis. 1982, 5, 47–55. [Google Scholar]
- St-Hilaire, S.; Ribble, C.; Traxler, G.; Davies, T.; Kent, M.L. Evidence for a carrier state of infectious hematopoietic necrosis virus in chinook salmon Oncorhynchus tshawytscha. Dis. Aquat. Organ. 2001, 46, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Neukirch, M. Demonstration of persistent viral haemorrhagic septicaemia (VHS) virus in rainbow trout after experimental waterborne infecton. J. Vet. Med. 1986, 33, 471–476. [Google Scholar]
- Hershberger, P.K.; Gregg, J.L.; Grady, C.A.; Taylor, L.; Winton, J.R. Chronic and persistent viral hemorrhagic septicemia virus infections in Pacific herring. Dis. Aquat. Organ. 2010, 93, 43–49. [Google Scholar]
- Amend, D.F. Detection and transmission of infectious hematopoietic necrosis virus in rainbow trout. J. Wildl. Dis. 1975, 11, 471–478. [Google Scholar]
- Engelking, H.M.; Harry, J.B.; Leong, J.C. Comparison of representative strains of infectious hematopoietic necrosis virus by serological neutralization and cross-protection assays. Appl. Environ. Microbiol. 1991, 57, 1372–1378. [Google Scholar]
- Lorenzen, N.; Lorenzen, E.; Einer-Jensen, K.; Heppell, J.; Wu, T.; Davis, H.L. Genetic vaccination of rainbow trout against viral haemorrhagic septicaemia virus: Small amounts of plasmid DNA protect against a heterologous serotype. Virus Res. 1999, 63, 19–25. [Google Scholar]
- Janeway, C.A.; Travers, P.; Walport, M.; Capra, D.J. Immunobiology, 4th ed; Garland Publishing: New York, NY, USA, 1999; p. 635. [Google Scholar]
- Lorenzen, N.; Olesen, N. Immunization with viral antigens: Viral haemorrhagic septicaemia. Dev. Biol. Stand. 1997, 90, 201–209. [Google Scholar]
- Winton, J.R. Immunization with viral antigens: Infectious haematopoietic necrosis. Dev. Biol. Stand. 1997, 90, 211–220. [Google Scholar]
- Lorenzen, N.; LaPatra, S.E. DNA vaccines for aquacultured fish. Revue Scientifique Et Technique-Office International Des Epizooties 2005, 24, 201–213. [Google Scholar]
- Lorenzen, N. Recombinant vaccines: Experimental and applied aspects. Fish Shellfish Immunol. 1999, 9, 361–365. [Google Scholar]
- Kurath, G. Overview of recent DNA vaccine development for fish. Dev. Biol. (Basel) 2005, 121, 201–213. [Google Scholar] [PubMed]
- Salonius, K.; Simard, N.; Harland, R.; Ulmer, J.B. The road to licensure of a DNA vaccine. Curr. Opin. Investig. Drugs 2007, 8, 635–641. [Google Scholar]
- Tonheim, T.C.; Bogwald, J.; Dalmo, R.A. What happens to the DNA vaccine in fish? A review of current knowledge. Fish Shellfish Immunol. 2008, 25, 1–18. [Google Scholar]
- Alonso, M.; Chiou, P.P.; Leong, J.A. Development of a suicidal DNA vaccine for infectious hematopoietic necrosis virus (IHNV). Fish Shellfish Immunol. 2011, 30, 815–823. [Google Scholar]
- Alonso, M.; Johnson, M.; Simon, B.; Leong, J.A. A fish specific expression vector containing the interferon regulatory factor 1A (IRF1A) promoter for genetic immunization of fish. Vaccine 2003, 21, 1591–1600. [Google Scholar]
- Anderson, E.D.; Mourich, D.V.; Fahrenkrug, S.C.; La Patra, S.E.; Shepard, J.; Leong, J.C. Genetic immunization of rainbow trout (Oncorhynchus mykiss) against infectious hematopoietic necrosis virus. Mol. Mar. Biol. Biotechnol. 1996, 5, 114–122. [Google Scholar] [PubMed]
- Lorenzen, E.; Lorenzen, E.; Einer-Jensen, K.; Heppell, J.; Wu, T.; Davis, H.L. Protective immunity to VHS in rainbow trout (Oncorhynchus mykiss Walbaum) following DNA vaccination. Fish Shellfish Immunol. 1998, 8, 261–270. [Google Scholar] [CrossRef]
- Traxler, G.S.; Anderson, E.; LaPatra, S.E.; Richard, J.; Shewmaker, B.; Kurath, G. Naked DNA vaccination of Atlantic salmon Salmo salar against IHNV. Dis. Aquat. Organ. 1999, 38, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Emmenegger, E.J.; Kurath, G. DNA vaccine protects ornamental koi (Cyprinus carpio koi) against North American spring viremia of carp virus. Vaccine 2008, 26, 6415–6421. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Iwahori, A.; Hirono, I.; Aoki, T. Development of a DNA vaccine against hirame rhabdovirus and analysis of the expression of immune-related genes after DNA vaccination. Fish Shellfish Immunol. 2004, 17, 367–374. [Google Scholar]
- Lorenzen, N.; Lorenzen, E.; Einer-Jensen, K.; Lapatra, S.E. DNA vaccines as a tool for analysing the protective immune response against rhabdoviruses in rainbow trout. Fish Shellfish Immunol. 2002, 12, 439–453. [Google Scholar]
- Kurath, G.; Purcell, M.K.; Garver, K.A. Fish rhabdovirus models for understanding host response to DNA vaccines. CAB Reviews 2007, 2, 1–12. [Google Scholar]
- Lorenzen, N.; Lorenzen, E.; Einer-Jensen, K.; La Patra, S.E. Immunity induced shortly after DNA vaccination of rainbow trout against rhabdoviruses protects against heterologous virus but not against bacterial pathogens. Dev. Comp. Immunol. 2002, 26, 173–179. [Google Scholar]
- LaPatra, S.E.; Corbeil, S.; Jones, G.R.; Shewmaker, W.D.; Lorenzen, N.; Anderson, E.D.; Kurath, G. Protection of rainbow trout against infectious hematopoietic necrosis virus four days after specific or semi-specific DNA vaccination. Vaccine 2001, 19, 4011–4019. [Google Scholar]
- Sommerset, I.; Lorenzen, E.; Lorenzen, N.; Bleie, H.; Nerland, A.H. A DNA vaccine directed against a rainbow trout rhabdovirus induces early protection against a nodavirus challenge in turbot. Vaccine 2003, 21, 4661–4667. [Google Scholar]
- Encinas, P.; Gomez-Sebastian, S.; Nunez, M.; Gomez-Casado, E.; Escribano, J.; Estepa, A.; Coll, J. Antibody recognition of the glycoprotein g of viral haemorrhagic septicemia virus (VHSV) purified in large amounts from insect larvae. BMC Res. Notes 2011, 4, 210. [Google Scholar]
- Adelmann, M.; Köllner, B.; Bergmann, S.M.; Fischer, U.; Lange, B.; Weitschies, W.; Enzmann, P.-J.; Fichtner, D. Development of an oral vaccine for immunisation of rainbow trout (Oncorhynchus mykiss) against viral haemorrhagic septicaemia. Vaccine 2008, 26, 837–844. [Google Scholar] [PubMed]
- Plant, K.P.; LaPatra, S.E. Advances in fish vaccine delivery. Dev. Comp. Immunol. 2011, 35, 1256–1262. [Google Scholar]
- Kawai, T.; Akira, S. Toll-like receptor and Rig-1 like receptor signaling. Ann. N. Y. Acad. Sci. 2008, 1143, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H.; Paludan, S.R. Reading the viral signature by Toll-like receptors and other pattern recognition receptors. J. Mol. Med. 2005, 83, 180–192. [Google Scholar]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 5598–5603. [Google Scholar]
- Georgel, P.; Jiang, Z.F.; Kunz, S.; Janssen, E.; Mols, J.; Hoebe, K.; Bahram, S.; Oldstone, M.B.; Beutler, B. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology 2007, 362, 304–313. [Google Scholar]
- Edelmann, K.H.; Richardson-Burns, S.; Alexopoulou, L.; Tyler, K.L.; Flavell, R.A.; Oldstone, M.B. Does Toll-like receptor 3 play a biological role in virus infections? Virology 2004, 322, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Faul, E.J.; Wanjalla, C.N.; Suthar, M.S.; Gale, M.; Wirblich, C.; Schnell, M.J. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 2010, 6, e1001016. [Google Scholar]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [PubMed]
- Samuel, C. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar]
- Ank, N.; West, H.; Paludan, S.R. IFN-lambda: Novel antiviral cytokines. J. Interferon Cytokine Res. 2006, 26, 373–379. [Google Scholar]
- Rieder, M.; Conzelmann, K.K. Rhabdovirus evasion of the interferon system. J. Interferon Cytokine Res. 2009, 29, 499–509. [Google Scholar]
- Komatsu, T.; Bi, Z.; Reiss, C.S. Interferon-gamma induced type I nitric oxide synthase activity inhibits viral replication in neurons. J. Neuroimmunol. 1996, 68, 101–108. [Google Scholar]
- D'Agostino, P.M.; Yang, J.; Reiss, C.S. Distinct mechanisms of inhibition of VSV replication in neurons mediated by type I and type II IFN. Virus Rev. Res. 2009, 14, 20–29. [Google Scholar]
- Sen, G.C. Viruses and interferons. Annu. Rev. Microbiol. 2001, 55, 255–281. [Google Scholar]
- Kalvakolanu, D.V. Alternate interferon signaling pathways. Pharmacol. Therapeut. 2003, 100, 1–29. [Google Scholar]
- van Boxel-Dezaire, A.H.; Rani, M.R.; Stark, G.R. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 2006, 25, 361–372. [Google Scholar]
- MacMicking, J.D. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 2004, 25, 601–609. [Google Scholar]
- Balachandran, S.; Roberts, P.C.; Brown, L.E.; Truong, H.; Pattnaik, A.K.; Archer, D.R.; Barber, G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 2000, 13, 129–141. [Google Scholar]
- Thomsen, A.R.; Nansen, A.; Andersen, C.; Johansen, J.; Marker, O.; Christensen, J.P. Cooperation of B cells and T cells is required for survival of mice infected with vesicular stomatitis virus. Int. Immunol. 1997, 9, 1757–1766. [Google Scholar]
- Freer, G.; Burkhart, C.; Ciernik, I.; Bachmann, M.F.; Hengartner, H.; Zinkernagel, R.M. Vesicular stomatitis virus Indiana glycoprotein as a T-cell-dependent and -independent antigen. J. Virol. 1994, 68, 3650–3655. [Google Scholar]
- Zou, J.; Bird, S.; Secombes, C. Antiviral sensing in teleost fish. Curr. Pharmaceut. Des. 2010, 16, 4185–4193. [Google Scholar]
- Palti, Y. Toll-like receptors in bony fish: From genomics to function. Dev. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar]
- Rebl, A.; Goldammer, T.; Seyfert, H.-M. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 2010, 134, 139–150. [Google Scholar]
- Huang, T.; Su, J.; Heng, J.; Dong, J.; Zhang, R.; Zhu, H. Identification and expression profiling analysis of grass carp Ctenopharyngodon idella LGP2 cDNA. Fish Shellfish Immunol. 2010, 29, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Huang, T.; Dong, J.; Heng, J.; Zhang, R.; Peng, L. Molecular cloning and immune responsive expression of MDA5 gene, a pivotal member of the RLR gene family from grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. 2010, 28, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S.; LeBerre, M.; Lamoureux, A.; Louise, Y.; Lauret, E.; Boudinot, P.; Bremont, M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 2009, 83, 7815–7827. [Google Scholar]
- Skjaeveland, I.; Iliev, D.B.; Strandskog, G.; Jorgensen, J.B. Identification and characterization of TLR8 and MyD88 homologs in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 2009, 33, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y.; Gahr, S.A.; Purcell, M.K.; Hadidi, S.; Rexroad, C.E., 3rd; Wiens, G.D. Identification, characterization and genetic mapping of TLR7, TLR8a1 and TLR8a2 genes in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2009, 34, 219–233. [Google Scholar] [PubMed]
- Rodriguez, M.F.; Wiens, G.; Purcell, M.K.; Palti, Y. Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics 2005, 57, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, A.; Oshiumi, H.; Tsujita, T.; Mitani, H.; Kasai, H.; Yoshimizu, M.; Matsumoto, M.; Seya, T. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 2008, 181, 3474–3485. [Google Scholar]
- Sullivan, C.; Charette, J.; Catchen, J.; Lage, C.R.; Giasson, G.; Postlethwait, J.H.; Millard, P.J.; Kim, C.H. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J. Immunol. 2009, 183, 5896–5908. [Google Scholar]
- Verjan, N.; Ooi, E.L.; Nochi, T.; Kondo, H.; Hirono, I.; Aoki, T.; Kiyono, H.; Yuki, Y. A soluble nonglycosylated recombinant infectous hematopoietic necrosis virus (IHNV) G-protein induces IFNs in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2008, 25, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Acosta, F.; Collet, B.; Lorenzen, N.; Ellis, A.E. Expression of the glycoprotein of viral haemorrhagic septicaemia virus (VHSV) on the surface of the fish cell line RTG-P1 induces type 1 interferon expression in neighbouring cells. Fish Shellfish Immunol. 2006, 21, 272–278. [Google Scholar]
- Chico, V.; Martinez-Lopez, A.; Ortega-Villaizan, M.; Falco, A.; Perez, L.; Coll, J.M.; Estepa, A. Pepscan mapping of viral hemorrhagic septicemia virus glycoprotein G major lineal determinants implicated in triggering host cell antiviral responses mediated by type I interferon. J. Virol. 2010, 84, 7140–7150. [Google Scholar]
- Chang, M.; Collet, B.; Nie, P.; Lester, K.; Campbell, S.; Secombes, C.J.; Zou, J. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in rainbow trout (Oncorhynchus mykiss). J. Virol. 2011, 85, 8403–8412. [Google Scholar] [PubMed]
- Altman, S.M.; Mellon, M.T.; Distel, D.L.; Kim, C.H. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 2003, 77, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Wilson, M.; Bengten, E.; Bryan, L.; Clem, L.W.; Miller, N.W.; Chinchar, V.G. Identification of a cDNA encoding channel catfish interferon. Dev. Comp. Immunol. 2004, 28, 97–111. [Google Scholar]
- Long, S.; Milev-Milovanovic, I.; Wilson, M.; Bengten, E.; Clem, L.W.; Miller, N.W.; Chinchar, V.G. Identification and expression analysis of cDNAs encoding channel catfish type I interferons. Fish Shellfish Immunol. 2006, 21, 42–59. [Google Scholar]
- Robertsen, B.; Bergan, V.; Rokenes, T.; Larsen, R.; Albuquerque, A. Atlantic salmon interferon genes: Cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 2003, 10, 601–612. [Google Scholar]
- Sun, B.; Robertsen, B.; Wang, Z.; Bin, L. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev. Comp. Immunol. 2008, 33, 547–558. [Google Scholar]
- Zou, J.; Tafalla, C.; Truckle, J.; Secombes, C. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 2007, 179, 3859–3871. [Google Scholar]
- Purcell, M.K.; Laing, K.J.; Woodson, J.; Thorgaard, G.; Hansen, J.D. Characterization of the interferon genes in homozygous rainbow trout reveals two novel genes, alternate splicing and differential regulation of duplicated genes. Fish Shellfish Immunol. 2009, 26, 293–304. [Google Scholar]
- Milev-Milovanovic, I.; Long, S.; Wilson, M.; Bengten, E.; Miller, N.W.; Chinchar, V.G. Identification and expression analysis of interferon gamma genes in channel catfish. Immunogenetics 2006, 58, 70–80. [Google Scholar]
- Zou, J.; Carrington, A.C.; Collet, B.; Dijkstra, J.M.; Yoshiura, Y.; Bols, N.; Secombes, C.J. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: The first Th1-type cytokine characterized functionally in fish. J. Immunol. 2005, 175, 2484–2494. [Google Scholar] [PubMed]
- Igawa, D.; Sakai, M.; Savan, R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol. Immunol. 2006, 43, 999–1009. [Google Scholar]
- Stein, C.; Caccamo, M.; Laird, G.; Leptin, M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007, 8, R251. [Google Scholar]
- Levraud, J.P.; Boudinot, P.; Colin, I.; Benmansour, A.; Peyrieras, N.; Herbomel, P.; Lutfalla, G. Identification of the zebrafish IFN receptor: Implications for the origin of the vertebrate IFN system. J. Immunol. 2007, 178, 4385–4394. [Google Scholar]
- Aggad, D.; Mazel, M.; Boudinot, P.; Mogensen, K.E.; Hamming, O.J.; Hartmann, R.; Kotenko, S.; Herbomel, P.; Lutfalla, G.; Levraud, J.P. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 2009, 183, 3924–3931. [Google Scholar]
- Zou, J.; Secombes, C.J. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 2011, 2011, 1376–1387. [Google Scholar]
- de Kinkelin, P.; Dorson, M. Interferon production in rainbow trout (Salmo gairdneri) experimentally infected with Egtved Virus. J. Gen. Virol. 1973, 19, 125–127. [Google Scholar] [CrossRef] [PubMed]
- de Kinkelin, P.; Dorson, M.; Hattenberger-Baudouy, A.M. Interferon synthesis in trout and carp after viral infection. Dev. Comp. Immunol. 1982, 2, 167–174. [Google Scholar]
- Eaton, W.D. Anti-viral activity in four species of salmonids following exposure to poly inosinic: Cytidylic acid. Dis. Aquat. Organ. 1990, 9, 193–198. [Google Scholar]
- Wang, L.; Zhang, H.X.; Zhang, J.H.; Chen, W.H.; Ruan, X.F.; Xia, C. In vitro effects of recombinant zebrafish IFN on spring viremia of carp virus and infectious hematopoietic necrosis virus. J. Interferon Cytokine Res. 2006, 26, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Ooi, E.L.; Verjan, N.; Haraguchi, I.; Oshima, T.; Kondo, H.; Hirono, I.; Aoki, T.; Kiyono, H.; Yuki, Y. Innate immunomodulation with recombinant interferon-alpha enhances resistance of rainbow trout (Oncorhynchus mykiss) to infectious hematopoietic necrosis virus. Dev. Comp. Immunol. 2008, 32, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- de Kinkelin, P.; Le Berre, M. Hematopoietic infectious necrosis of Salmonidae: Production of circulating interferon after experimental infection of the rainbow trout (Salmo gairdneri). C. R. Acad. Sci. Hebd. Seances Acad. Sci. D. 1974, 279, 445–448. [Google Scholar] [PubMed]
- Trobridge, G.D.; Chiou, P.P.; Leong, J.C. Cloning of the rainbow trout (Oncorhynchus mykiss) Mx2 and Mx3 cDNAs and characterization of trout Mx protein expression in salmon cells. J. Virol. 1997, 71, 5304–5311. [Google Scholar] [PubMed]
- O'Farrell, C.; Vaghefi, N.; Cantonnet, M.; Buteau, B.; Boudinot, P.; Benmansour, A. Survey of transcript expression in rainbow trout leukocytes reveals a major contribution of interferon-responsive genes in the early response to a rhabdovirus infection. J. Virol. 2002, 76, 8040–8049. [Google Scholar]
- Purcell, M.K.; Kurath, G.; Garver, K.A.; Herwig, R.P.; Winton, J.R. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immunol. 2004, 17, 447–462. [Google Scholar]
- Collet, B.; Boudinot, P.; Benmansour, A.; Secombes, C.J. An Mx1 promoter-reporter system to study interferon pathways in rainbow trout. Dev. Comp. Immunol. 2004, 28, 793–801. [Google Scholar]
- Lopez-Munoz, A.; Roca, F.J.; Meseguer, J.; Mulero, V. New insights into the evolution of IFNs: Zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J. Immunol. 2009, 182, 3440–3449. [Google Scholar]
- Chaves-Pozo, E.; Zou, J.; Secombes, C.J.; Cuesta, A.; Tafalla, C. The rainbow trout (Oncorhynchus mykiss) interferon response in the ovary. Mol. Immunol. 2010, 47, 1757–1764. [Google Scholar] [PubMed]
- Martin, S.A.; Taggart, J.B.; Seear, P.; Bron, J.E.; Talbot, R.; Teale, A.J.; Sweeney, G.E.; Hoyheim, B.; Houlihan, D.F.; Tocher, D.R.; et al. Interferon type I and type II responses in an Atlantic salmon (Salmo salar) SHK-1 cell line by the salmon TRAITS/SGP microarray. Physiol. Genom. 2007, 32, 33–44. [Google Scholar] [CrossRef]
- Sun, B.; Skjaeveland, I.; Svingerud, T.; Zou, J.; Jorgensen, J.; Robertsen, B. Antiviral activity of salmonid gamma interferon against infectious pancreatic necrosis virus and salmonid alphavirus and its dependency on type I interferon. J. Virol. 2011, 85, 9188–9198. [Google Scholar]
- Verrier, E.R.; Langevin, C.; Benmansour, A.; Boudinot, P. Early antivirial response and virus-induced genes in fish. Dev. Comp. Immunol. 2011, 35, 1204–1214. [Google Scholar]
- Byon, J.Y.; Ohira, T.; Hirono, I.; Aoki, T. Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceus) following DNA vaccination. Fish Shellfish Immunol. 2005, 18, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Byon, J.Y.; Ohira, T.; Hirono, I.; Aoki, T. Comparative immune responses in Japanese flounder, Paralichthys olivaceus after vaccination with viral hemorrhagic septicemia virus (VHSV) recombinant glycoprotein and DNA vaccine using a microarray analysis. Vaccine 2006, 24, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Nichols, K.M.; Winton, J.R.; Kurath, G.; Thorgaard, G.H.; Wheeler, P.; Hansen, J.D.; Herwig, R.P.; Park, L.K. Comprehensive gene expression profiling following DNA vaccination of rainbow trout against Infectious hematopoietic necrosis virus. Mol. Immunol. 2006, 43, 2089–2106. [Google Scholar] [CrossRef] [PubMed]
- Peñaranda, M.M.D.; Purcell, M.K.; Kurath, G. Differential virulence mechanisms of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J. Gen. Virol. 2009, 90, 2172–2182. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Garver, K.A.; Conway, C.M.; Elliott, D.G.; Kurath, G. Infectious hematopoietic necrosis virus genogroup-specfic virulence mechanisms in sockeye salmon (Oncorhynchus nerka) from Redfish Lake Idaho. J. Fish Dis. 2009, 32, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J.; Bearzotti-le Berre, M.; De Kinkelin, P. Viral haemorrhagic septicaemia in rainbow trout: Attempt to relate interferon production, antibody synthesis and structure of the virus with the mechanism of virulence. Annalles Institute Pasteur 1985, 136, 13–26. [Google Scholar]
- Park, J.W.; Moon, C.H.; Wargo, A.W.; Purcell, M.K.; Kurath, G. Differential growth of U and M type infectious hematopoietic necrosis virus in a rainbow trout-derived cell line, RTG-2. J. Fish Dis. 2010, 33, 583–591. [Google Scholar]
- Wargo, A.R.; Garver, K.A.; Kurath, G. Virulence correlates with fitness in vivo for two M group genotypes of Infectious hematopoietic necrosis virus (IHNV). Virology 2010, 404, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wargo, A.R.; Kurath, G. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J. Virol. 2011, 85, 3959–3967. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Lapatra, S.E.; Woodson, J.C.; Kurath, G.; Winton, J.R. Early viral replication and induced or constitutive immunity in rainbow trout families with differential resistance to Infectious hematopoietic necrosis virus (IHNV). Fish Shellfish Immunol. 2009, 28, 98–105. [Google Scholar]
- Dorson, M.; Torhy, C. Viral haemorrhagic septicaemia virus replication in external tissue excised from rainbow trout, Oncorhynchus mykiss (Walbaum), and hybrids of different susceptibilities. J. Fish Dis. 1993, 16, 403–408. [Google Scholar] [CrossRef]
- Quillet, E.; Dorson, M.; Aubard, G.; Torhy, C. In vitro viral haemorrhagic septicemia virus replication in excised fins of rainbow trout: Correlation with resistance to waterborne challenge and genetic variation. Dis. Aquat. Organ. 2001, 45, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Quillet, E.; Dorson, M.; Aubard, G.; Torhy, C. In vitro assay to select rainbow trout with variable resistance/susceptibility to viral haemorrhagic septicaemia virus. Dis. Aquat. Organ. 2007, 76, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Harmache, A.; LeBerre, M.; Droineau, S.; Giovannini, M.; Bremont, M. Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus. J. Virol. 2006, 80, 3655–3659. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, N.; LaPatra, S.E. Immunity to rhabdoviruses in rainbow trout: The antibody response. Fish Shellfish Immunol. 1999, 9, 345–360. [Google Scholar]
- LaPatra, S.E.; Turner, T.; Lauda, K.A.; Jones, G.R.; Walker, S. Characterization of the humoral response of rainbow trout to infectious hematopoietic necrosis virus. J. Aquat. Anim. Health 1993, 5, 165–171. [Google Scholar]
- LaPatra, S.E.; Lauda, K.A.; Jones, G.R.; Walker, S.; Shewmaker, W.D. Development of passive immunotherapy for control of infectious haematopoietic necrosis. Dis. Aquat. Organ. 1994, 20, 1–6. [Google Scholar]
- Hershberger, P.K.; Gregg, J.L.; Grady, C.A.; LaPatra, S.E.; Winton, J.R. Passive immunization of Pacific herring Clupea pallasii against viral hemorrhagic septicemia. J. Aquat. Anim. Health 2011, 23, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, N.; Olesen, N.J.; Koch, C. Immunity to VHS virus in rainbow trout. Aquaculture 1999, 172, 41–61. [Google Scholar]
- Kurath, G.; Garver, K.A.; LaPatra, S.E.; Purcell, M.K. Resistance and protective immunity in Redfish Lake sockeye salmon exposed to M type infectious hematopoietic necrosis virus (IHNV). J. Aquat. Anim. Health 2010, 22, 129–139. [Google Scholar]
- McLauchlan, P.E.; Collet, B.; Ingerslev, E.; Secombes, C.J.; Lorenzen, N.; Ellis, A.E. DNA vaccination against viral haemorrhagic septicaemia (VHS) in rainbow trout: Size, dose, route of injections and duration of protection-early protection correlates with Mx expression. Fish Shellfish Immunol. 2003, 15, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, S.; LaPatra, S.E.; Anderson, E.D.; Kurath, G. Nanogram quantities of a DNA vaccine protect rainbow trout fry against heterologous virus strains of infectious hematopoietic necrosis virus. Vaccine 2000, 18, 2817–2824. [Google Scholar]
- LaPatra, S.E.; Corbeil, S.; Jones, G.R.; Shewmaker, W.D.; Kurath, G. The dose-dependent effect on protection and humoral response to a DNA vaccine against IHN virus in subyearling rainbow trout. J. Aquat. Anim. Health 2000, 12, 181–188. [Google Scholar]
- Garver, K.A.; LaPatra, S.E.; Kurath, G. Efficacy of an infectious hematopoietic necrosis (IHN) virus DNA vaccine in Chinook Oncorhynchus tshawytscha and sockeye O. nerka salmon. Dis. Aquat. Organ. 2005, 64, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Boudinot, P.; Blanco, M.; de Kinkelin, P.; Benmansour, A. Combined DNA immunization with the glycoprotein gene of viral hemorrhagic septicemia virus and infectious hematopoeitic necrosis virus induces double-specific immunity and nonspecific responses in rainbow trout. Virology 1998, 249, 297–306. [Google Scholar]
- Kurath, G.; Garver, K.A.; Corbeil, S.; Elliott, D.G.; Anderson, E.D.; LaPatra, S.E. Protective immunity and lack of histopathological damage two years after DNA vaccination against infectious hematopoietic necrosis virus in trout. Vaccine 2006, 16, 345–354. [Google Scholar]
- Li, J.; Barreda, D.R.; Zhang, Y.A.; Boshra, H.; Gelman, A.E.; Lapatra, S.; Tort, L.; Sunyer, J.O. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 2006, 7, 1116–1124. [Google Scholar]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 6919–6924. [Google Scholar]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar]
- Hordvik, I.; Berven, F.S.; Solem, S.T.; Hatten, F.; Endresen, C. Analysis of two IgM isotypes in Atlantic salmon and brown trout. Mol. Immunol. 2002, 39, 313–321. [Google Scholar]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar]
- DeLuca, D.; Wilson, M.; Warr, G.W. Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur. J. Immunol. 1983, 13, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Tsujikura, M.; Ichiki, S.; Vo, T.K.; Somamoto, T. The complement system in teleost fish: Progress of post-homolog-hunting researches. Dev. Comp. Immunol. 2011, 35, 1296–1308. [Google Scholar]
- Laing, K.J.; Hansen, J.D. Fish T cells: Recent advances through genomics. Dev. Comp. Immunol. 2011, 35, 1282–1295. [Google Scholar]
- Miller, K.; Traxler, G.; Kaukinen, K.; Li, S.; Richard, J.; Ginther, N. Salmonid host response to infectious hematopoietic necrosis (IHN) virus: Cellular receptors, viral control, and novel pathways of defense. Aquaculture 2007, 272, S217–S237. [Google Scholar] [CrossRef]
- Boudinot, P.; Bernard, D.; Boubekeur, S.; Thoulouze, M.-I.; Bremont, M.; Benmansour, A. The glycoprotein of a fish rhabdovirus profiles the virus-specific T-cell repertoire in rainbow trout. J. Gen. Virol. 2004, 85, 3099–3108. [Google Scholar]
- Boudinot, P.; Boubekeur, S.; Benmansour, A. Rhabdovirus infection induces public and private T cell responses in teleost fish. J. Immunol. 2001, 167, 6202–6209. [Google Scholar]
- Takizawa, F.; Dijkstra, J.M.; Kotterba, P.; Korytar, T.; Kock, H.; Kollner, B.; Jaureguiberry, B.; Nakanishi, T.; Fischer, U. The expression of CD8 alpha discriminates distinct T cell subsets in teleost fish. Dev. Comp. Immunol. 2011, 35, 752–763. [Google Scholar]
- Nakanishi, T.; Toda, H.; Shibasaki, Y.; Somamoto, T. Cytotoxic T cells in teleost fish. Dev. Comp. Immunol. 2011, 35, 1317–1323. [Google Scholar]
- Fischer, U.; Utke, K.; Somamoto, T.; Kollner, B.; Ototake, M.; Nakanishi, T. Cytotoxic activities of fish leucocytes. Fish Shellfish Immunol. 2006, 20, 209–226. [Google Scholar]
- Evans, D.E.; Jaso-Friedman, L. Nonspecific cytotoxic cells as effectors of immunity in fish. Annu. Rev. Fish Dis. 1992, 2, 109–121. [Google Scholar]
- Shen, L.L.; Stuge, T.B.; Evenhuis, J.P.; Bengten, E.; Wilson, M.; Chinchar, V.G.; Clem, L.W.; Miller, N.W. Channel catfish NK-like cells are armed with IgM via a putative FcmuR. Dev. Comp. Immunol. 2003, 27, 699–714. [Google Scholar]
- Shen, L.; Stuge, T.B.; Bengten, E.; Wilson, M.; Chinchar, V.G.; Naftel, J.P.; Bernanke, J.M.; Clem, L.W.; Miller, N.W. Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus). Dev. Comp. Immunol. 2004, 28, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Kurata, O.; Okamoto, N.; Ikeda, Y. Neutrophilic granulocytes in carp, Cyprinus carpio, possess a spontaneous cytotoxic activity. Dev. Comp. Immunol. 1995, 19, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Litman, G.W. The phylogenetic origins of natural killer receptors and recognition: Relationships, possibilities, and realities. Immunogenetics 2011, 63, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Kollner, B.; Aoyagi, K.; Sawamoto, Y.; Kuroda, A.; Ototake, M.; Nakanishi, T.; Fischer, U. The rainbow trout classical MHC class I molecule Onmy-UBA*501 is expressed in similar cell types as mammalian classical MHC class I molecules. Fish Shellfish Immunol. 2003, 14, 1–23. [Google Scholar]
- Utke, K.; Bergmann, S.; Lorenzen, N.; Kollner, B.; Ototake, M.; Fischer, U. Cell-mediated cytotoxicity in rainbow trout, Oncorhynchus mykiss, infected with viral haemorrhagic septicaemia virus. Fish Shellfish Immunol. 2007, 22, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Somamoto, T.; Nakanishi, T.; Okamoto, N. Role of specific cell-mediated cytotoxicity in protecting fish from viral infections. Virology 2002, 297, 120–127. [Google Scholar]
- Le Morvan, C.; Troutaud, D.; Deschaux, P. Differential effects of temperature on specific and nonspecific immune defenses in fish. J. Exp. Biol. 1998, 201, 165–168. [Google Scholar]
- Fijan, N. Spring viraemia of carp and other viral diseases and agents of warm-water fish. In Fish Diseases and Disorders; Woo, P.T.K., Bruno, D.W., Eds.; CAB International: Wallingford, UK, 1999; Volume 3, pp. 177–244. [Google Scholar]
- Wolf, K. Viral hemorrhagic septicemia. In Fish Viruses and Fish Viral Diseases; Cornell University Press: Ithaca, NY, USA, 1988; pp. 217–249. [Google Scholar]
- Amend, D.F. Prevention and control of viral diseases of salmonids. J. Fish. Res. Board Can. 1976, 33, 1059–1066. [Google Scholar]
- Goodwin, A.E.; Merry, G.E. Mortality and carrier status of bluegills exposed to viral hemorrhagic septicemia virus genotype IVb at different temperatures. J. Aquat. Anim. Health 2011, 23, 85–91. [Google Scholar]
- Amend, D.F. Control of infectious hematopoietic necrosis virus disease by elevating the water temperature. J. Fish. Res. Board Can. 1970, 27, 265–270. [Google Scholar]
- Sano, M.; Ito, M.T.; Nakayasu, C.; Kurita, J. Effect of water temperature shifting on mortality of Japanese flounder Paralichthys olivaceus experimentally infected with viral hemorrhagic septicemia virus. Aquaculture 2009, 286, 254–258. [Google Scholar] [CrossRef]
- LaPatra, S.E.; Batts, W.N.; Overturf, K.; Jones, G.R.; Shewmaker, W.D.; Winton, J.R. Negligible risk associated with the movement of processed rainbow trout, Oncorhynchus mykiss (Walbaum), from an infectious haematopoietic necrosis virus (IHNV) endemic area. J. Fish Dis. 2001, 24, 399–408. [Google Scholar] [CrossRef]
- Lorenzen, E.; Einer-Jensen, K.; Rasmussen, J.S.; Kjaer, T.E.; Collet, B.; Secombes, C.J.; Lorenzen, N. The protective mechanisms induced by a fish rhabdovirus DNA vaccine depend on temperature. Vaccine 2009, 27, 3870–3880. [Google Scholar]
- Alcorn, S.W.; Murray, A.L.; Pascho, R.J. Effects of rearing temperature on immune functions in sockeye salmon (Oncorhynchus nerka). Fish Shellfish Immunol. 2002, 12, 303–334. [Google Scholar] [CrossRef] [PubMed]
- Utke, K.; Kock, H.; Schuetze, H.; Bergmann, S.M.; Lorenzen, N.; Einer-Jensen, K.; Kollner, B.; Dalmo, R.A.; Vesely, T.; Ototake, M.; et al. Cell-mediated immune responses in rainbow trout after DNA immunization against the viral hemorrhagic septicemia virus. Dev. Comp. Immunol. 2008, 32, 239–252. [Google Scholar] [PubMed]
- Sealey, W.M.; Barrows, F.T.; Johansen, K.A.; Overturf, K.; LaPatra, S.E.; Hardy, R.W. Evaluation of the ability of partially autolyzed yeast and Grobiotic-A to improve disease resistance in rainbow trout. N. Am. J. Aquaculture 2007, 69, 400–406. [Google Scholar]
- Beaulaurier, J.; Bickford, N.; Gregg, J.L.; Grady, C.A.; Gannam, A.; Winton, J.R.; Hershberger, P.K. Susceptibility of Pacific herring Clupea pallasii to viral hemorrhagic septicemia (VHS) is influenced by diet. J. Aquat. Anim. Health 2012, in press. [Google Scholar]
- Lyles, D.S. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 2000, 64, 709–724. [Google Scholar]
- Ahmed, M.; Lyles, D.S. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and II. J. Virol. 1998, 72, 8413–8419. [Google Scholar]
- Rieder, M.; Brzozka, K.; Pfaller, C.K.; Cox, J.H.; Stitz, L.; Conzelmann, K.K. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: Inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J. Virol. 2011, 85, 842–852. [Google Scholar]
- Blondel, D.; Regad, T.; Poisson, N.; Pavie, B.; Harper, F.; Pandolfi, P.P.; De The, H.; Chelbi-Alix, M.K. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 2002, 21, 7957–7970. [Google Scholar]
- Petersen, J.M.; Her, L.S.; Varvel, V.; Lund, E.; Dahlberg, J.E. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Biol. Cell 2000, 20, 8590–8601. [Google Scholar]
- Connor, J.H.; Lyles, D.S. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J. Biol. Chem. 2005, 280, 13512–13519. [Google Scholar] [PubMed]
- Benedict, C.A.; Norris, P.S.; Ware, C.F. To kill or be killed: Viral evasion of apoptosis. Nat. Immunol. 2002, 3, 1013–1018. [Google Scholar]
- Gaddy, D.F.; Lyles, D.S. Vesicular stomatitis viruses expressing wild-type or mutant M proteins activate apoptosis through distinct pathways. J. Virol. 2005, 79, 4170–4179. [Google Scholar]
- Kopecky, S.A.; Willingham, M.C.; Lyles, D.S. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 2001, 75, 12169–12181. [Google Scholar]
- Kopecky, S.A.; Lyles, D.S. Contrasting effects of matrix protein on apoptosis in HeLa and BHK cells infected with vesicular stomatitis virus are due to inhibition of host gene expression. J. Virol. 2003, 77, 4658–4669. [Google Scholar]
- Koyama, A.H. Induction of apoptotic DNA fragmentation by the infection of vesicular stomatitis virus. Virus Res. 1995, 37, 285–290. [Google Scholar]
- Frank, S.A. Immunology and Evolution of Infectious Disease; Princeton University Press: Princeton, NJ, USA, 2002; p. 348. [Google Scholar]
- Benmansour, A.; Leblois, H.; Coulon, P.; Tuffereau, C.; Gaudin, Y.; Flamand, A.; Lafay, F. Antigenicity of rabies virus glycoprotein. J. Virol. 1991, 65, 4198–4203. [Google Scholar]
- Luo, L.H.; Li, Y.; Snyder, R.M.; Wagner, R.R. Point mutations in glycoprotein gene of vesicular stomatitis virus (New Jersey serotype) selected by resistance to neutralization by epitope-specific monoclonal antibodies. Virology 1988, 163, 341–348. [Google Scholar]
- Bilsel, P.A.; Nichol, S.T. Polymerase errors accumulating during natural evolution of the glycoprotein gene of vesicular stomatitis virus Indiana serotype isolates. J. Virol. 1990, 64, 4873–4883. [Google Scholar]
- Luo, L.Z.; Li, Y.; Snyder, R.M.; Wagner, R.R. Spontaneous mutations leading to antigenic variations in the glycoproteins of vesicular stomatitis virus field isolates. Virology 1990, 174, 70–78. [Google Scholar]
- Sloan, S.E.; Hanlon, C.; Weldon, W.; Niezgoda, M.; Blanton, J.; Self, J.; Rowley, K.J.; Mandell, R.B.; Babcock, G.J.; Thomas, W.D., Jr.; et al. Identification and characterization of a human monoclonal antibody that potently neutralizes a broad panel of rabies virus isolates. Vaccine 2007, 25, 2800–2810. [Google Scholar]
- Chiou, P.P.; Kim, C.H.; Ormonde, P.; Leong, J.A. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. J. Virol. 2000, 74, 7619–7627. [Google Scholar]
- Ammayappan, A.; Vakharia, V.N. Nonvirion protein of novirhabdovirus suppresses apoptosis at the early stage of virus infection. J. Virol. 2011, 85, 8393–8402. [Google Scholar]
- Johnson, M.C.; Simon, B.E.; Kim, C.H.; Leong, J.-A.C. Production of recombinant snakehead rhabdovirus: The NV protein is not required for viral replication. J. Virol. 2000, 74, 2343–2350. [Google Scholar]
- Biacchesi, S.; Thoulouze, M.I.; Bearzotti, M.; Yu, Y.X.; Bremont, M. Recovery of NV knockout infectious hematopoietic necrosis virus expressing foreign genes. J. Virol. 2000, 74, 11247–11253. [Google Scholar]
- Fijan, N.; Sulimanovic, D.; Bearzotti, M.; Muzinic, D.; Zwillenberg, L.; Chilmonczyk, S.; Vautherot, J.F.; de Kinkelin, P. Some properties of the epithelioma papulosum cyprini (EPC) cell line from carp Cyprinus carpio. Ann. Virol. 1983, 134E, 207–220. [Google Scholar]
- Winton, J.; Batts, W.; deKinkelin, P.; LeBerre, M.; Bremont, M.; Fijan, N. Current lineages of the epithelioma papulosum cyprini (EPC) cell line are contaminated with fathead minnow, Pimephales promelas, cells. J. Fish Dis. 2010, 33, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Alonso, M.; Kim, C.H.; Johnson, M.C.; Pressley, M.; Leong, J.-A. The NV gene of snakehead rhabdovirus (SHRV) is not required for pathogenesis, and a heterologous glycoprotein can be incorporated into the SHRV envelope. J. Virol. 2004, 78, 5875–5882. [Google Scholar]
- Thoulouze, M.-I.; Bouguyon, E.; Carpentier, C.; Bremont, M. Essential Role of the NV Protein of Novirhabdovirus for Pathogenicity in Rainbow Trout. J. Virol. 2004, 78, 4098–4107. [Google Scholar]
- Ammayappan, A.; Kurath, G.; Thompson, T.M.; Vakharia, V.N. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: The NV gene is required for pathogenicity. Mar. Biotechnol. 2011, 13, 672–683. [Google Scholar]
- Choi, M.K.; Moon, C.H.; Ko, M.S.; Lee, U.H.; Cho, W.J.; Cha, S.J.; Do, J.W.; Heo, G.J.; Jeong, S.G.; Hahm, Y.S.; et al. A Nuclear Localization of the Infectious Haematopoietic Necrosis Virus NV Protein Is Necessary for Optimal Viral Growth. PLoS One 2011, 6, e22362. [Google Scholar] [PubMed]
- Smail, D.A. Viral haemorrhagic septicaemia. In Fish Diseases and Disorders: Viral, Bacterial, and Fungal Infections; Woo, P.T.K., Bruno, D.W., Eds.; CAB International: Wallingford, UK, 1999; Volume 3, pp. 123–147. [Google Scholar]
- Huang, C.; Chien, M.S.; Landolt, M.; Batts, W.; Winton, J. Mapping the neutralizing epitopes on the glycoprotein of infectious haematopoietic necrosis virus, a fish rhabdovirus. J. Gen. Virol. 1996, 77, 3033–3040. [Google Scholar]
- Nichol, S.T.; Rowe, J.E.; Winton, J.R. Molecular epizootiology and evolution of the glycoprotein and non-virion protein genes of infectious hematopoietic necrosis virus, a fish rhabdovirus. Virus Res. 1995, 38, 159–173. [Google Scholar]
- Kurath, G.; Garver, K.A.; Troyer, R.M.; Emmenegger, E.J.; Einer-Jensen, K.; Anderson, E.D. Phylogeography of infectious haematopoietic necrosis virus in North America. J. Gen. Virol. 2003, 84, 803–814. [Google Scholar]
- Troyer, R.M.; LaPatra, S.E.; Kurath, G. Genetic analyses reveal unusually high diversity of infectious haematopoietic necrosis virus in rainbow trout aquaculture. J. Gen. Virol. 2000, 81, 2823–2832. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Purcell, M.K.; Laing, K.J.; Winton, J.R. Immunity to Fish Rhabdoviruses. Viruses 2012, 4, 140-166. https://doi.org/10.3390/v4010140
Purcell MK, Laing KJ, Winton JR. Immunity to Fish Rhabdoviruses. Viruses. 2012; 4(1):140-166. https://doi.org/10.3390/v4010140
Chicago/Turabian StylePurcell, Maureen K., Kerry J. Laing, and James R. Winton. 2012. "Immunity to Fish Rhabdoviruses" Viruses 4, no. 1: 140-166. https://doi.org/10.3390/v4010140



