Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and RNA Preparations
2.2. Bisulfite Sequencing Protocols
2.3. RT-PCR, Cloning and Sequencing
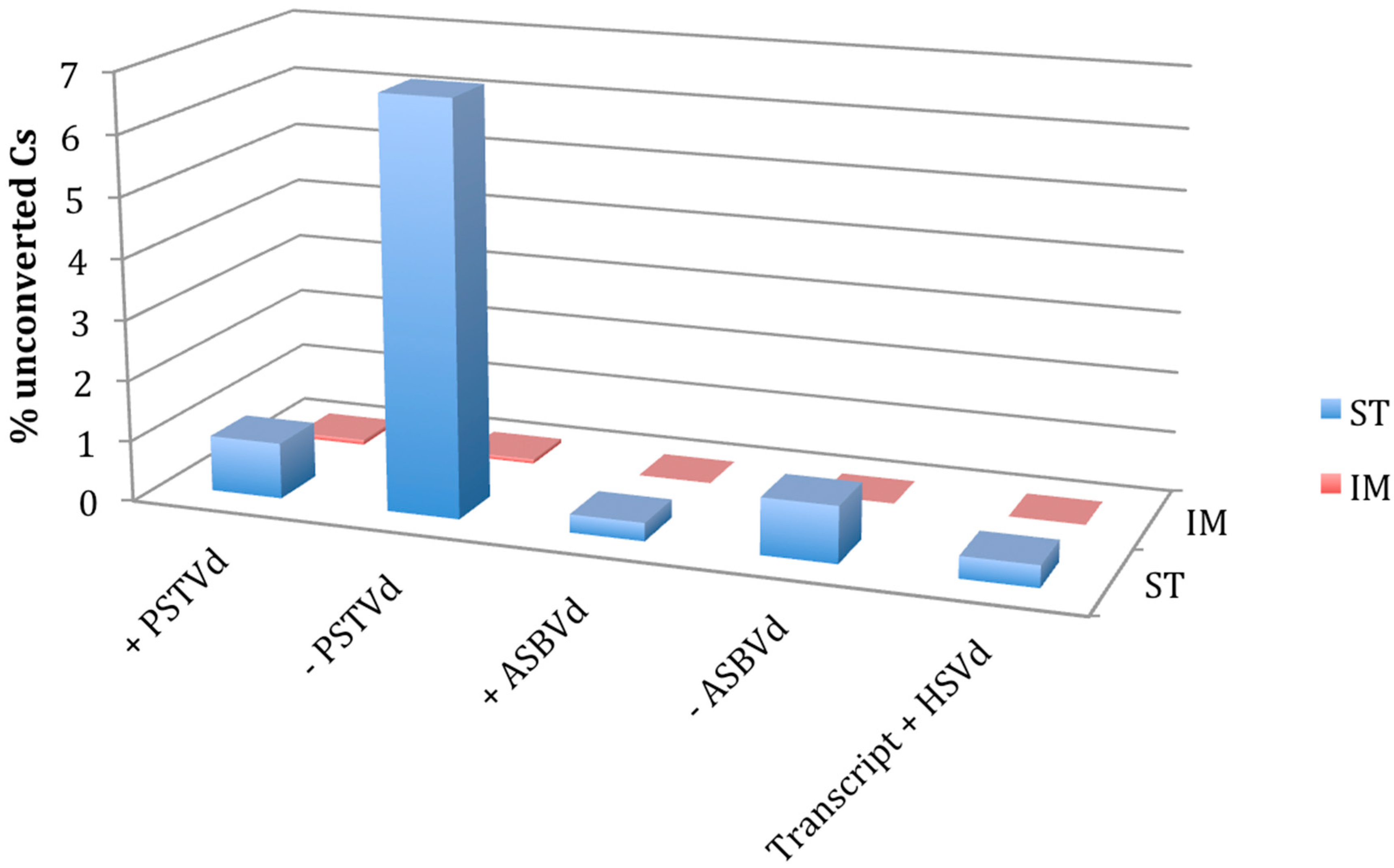
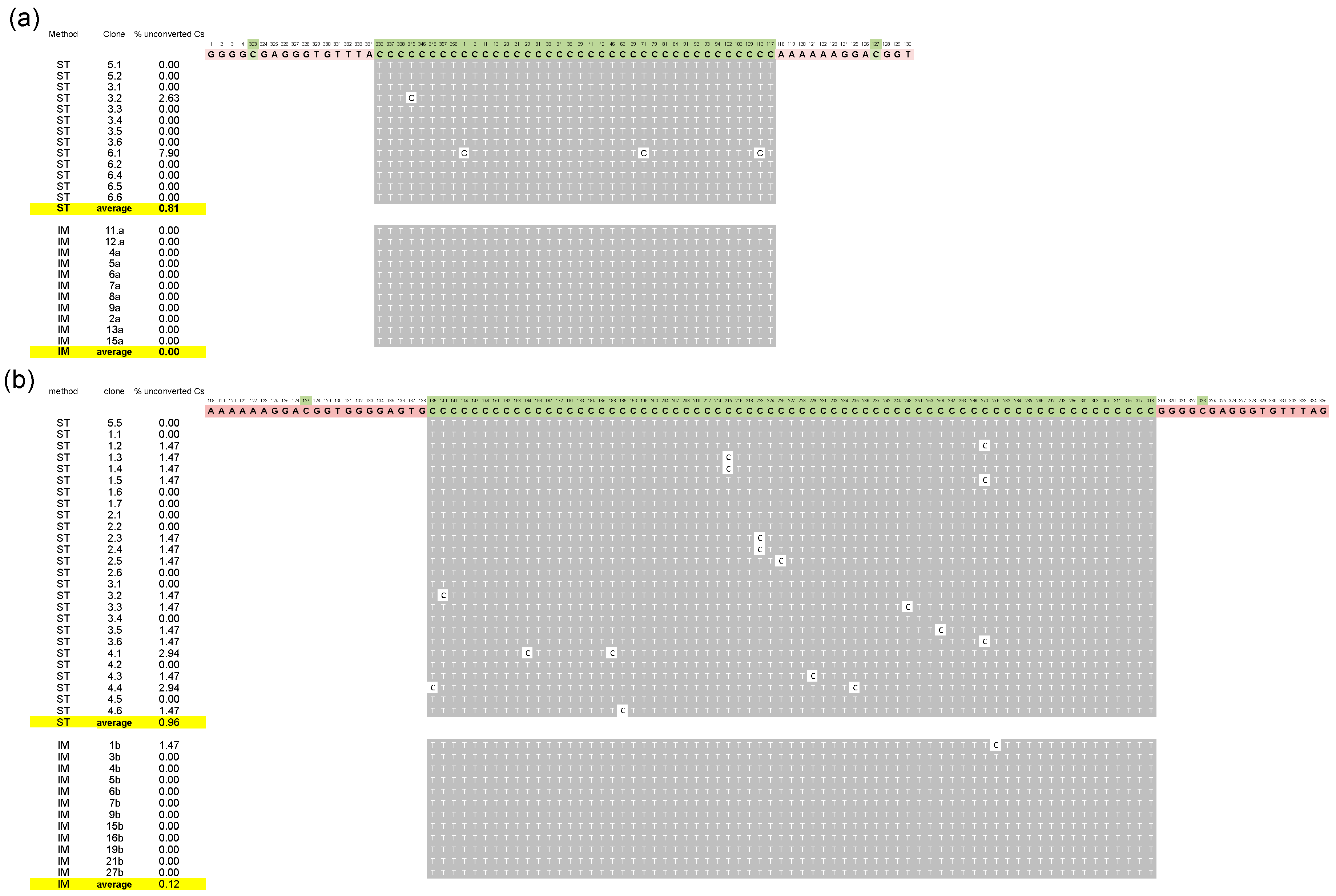
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sharp, P.A. The centrality of RNA. Cell 2009, 136, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Lyko, F.; Helm, M. 5-methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010, 38, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Pan, T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011, 44, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ma, P.; Liu, Y.; Li, W.; Shu, Y. Multiple functions of m(6)A RNA methylation in cancer. J. Hematol. Oncol. 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef]
- Grosjean, H. Modification and editing of RNA: Historical overview and important facts to remember. In Fine-Tuning of RNA Functions by Modification and Editing; Grosjean, H., Ed.; Springer: Berlin, Germany, 2005; pp. 1–22. [Google Scholar]
- Song, X.; Nazar, R.N. Modification of rRNA as a ‘quality control mechanism’ in ribosome biogenesis. FEBS Lett. 2002, 523, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef]
- Schaefer, M.; Lyko, F. Solving the Dnmt2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitzm, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Cui, X.; Liang, Z.; Shen, L.; Zhang, Q.; Bao, S.; Geng, Y.; Zhang, B.; Leo, V.; Vardy, L.A.; Lu, T.; et al. 5-Methylcytosine RNA methylation in Arabidopsis Thaliana. Mol. Plant 2017, 10, 1387–1399. [Google Scholar] [CrossRef]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals m5C within Archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef]
- Schaefer, M.; Pollex, T.; Hanna, K.; Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009, 37, e12. [Google Scholar] [CrossRef] [PubMed]
- Pollex, T.; Hanna, K.; Schaefer, M. Detection of cytosine methylation in RNA using bisulfite sequencing. Cold Spring Harb. Protoc. 2010, 10, pdb.prot5505. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amort, T.; Soulière, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Wörle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef]
- Tsagris, E.M.; Martínez de Alba, A.E.; Gozmanova, M.; Kalantidis, K. Viroids. Cell Microbiol. 2008, 10, 2168–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, R.; Minoia, S.; Carbonell, A.; Gisel, A.; Delgado, S.; López-Carrasco, A.; Navarro, B.; Di Serio, F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015, 209, 136–145. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Viroids: How to infect a host and cause disease without encoding proteins. Biochimie 2012, 94, 1474–1480. [Google Scholar] [CrossRef]
- Flores, R.; Serra, P.; Minoia, S.; Di Serio, F.; Navarro, B. Viroids: From genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbiol. 2012, 3, 217. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. Viroids: Self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip. Rev. RNA 2010, 1, 362–375. [Google Scholar] [CrossRef]
- Di Serio, F.; Flores, R.; Verhoeven, J.T.; Li, S.F.; Pallás, V.; Randles, J.W.; Sano, T.; Vidalakis, G.; Owens, R.A. Current status of viroid taxonomy. Arch. Virol. 2014, 159, 3467–3478. [Google Scholar] [CrossRef] [Green Version]
- Di Serio, F.; Li, S.F.; Matoušek, J.; Owens, R.A.; Pallás, V.; Randles, J.W.; Sano, T.; Verhoeven, J.T.J.; Vidalakis, G.; Flores, R. ICTV Virus Taxonomy Profile: Avsunviroidae. J. Gen. Virol. 2018, 99, 611–612. [Google Scholar] [CrossRef] [Green Version]
- Domdey, H.; Jank, P.; Sänger, L.; Gross, H.J. Studies on the primary and secondary structure of potato spindle tuber viroid: Products of digestion with ribonuclease A and ribonuclease T1, and modification with bisulfite. Nucleic Acids Res. 1978, 5, 1221–1236. [Google Scholar] [CrossRef]
- Goddard, J.P.; Schulman, L.H. Conversion of exposed cytidine residues to uridine residues in Escherichia coli formylmethionine transfer ribonucleic acid. J. Biol. Chem. 1972, 247, 3864–3867. [Google Scholar]
- Goddard, J.P.; Maden, B.E. Reaction of HeLa cell methyl-labelled 28S ribosomal RNA with sodium bisulfite: A conformational probe for methylated sequences. Nucleic Acids Res. 1976, 3, 431–440. [Google Scholar] [CrossRef]
- Pallas, V.; Navarro, A.; Flores, R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J. Gen. Virol. 1987, 68, 3201–3205. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Takamatsu, N.; Meshi, T.; Okada, Y. Hop stunt viroid: Molecular cloning and nucleotide sequence of the complete cDNA copy. Nucleic Acids Res. 1983, 11, 6185–6197. [Google Scholar] [CrossRef] [PubMed]
- Duran-Vila, N.; Elena, S.F.; Daròs, J.A.; Flores, R. Structure and Evolution of Viroids. In Origin and Evolution of Viruses, 2nd ed.; Domingo, E., Parrish, C.R., Holland, J.J., Eds.; Academic Press: Cambridge, MA, USA, 2008; Chapter 2; pp. 43–64. ISBN 978-0-12-374153-0. [Google Scholar]
- Trixl, L.; Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. WIREs RNA 2019, 10, e1510. [Google Scholar] [CrossRef] [PubMed]
- García-Vílchez, R.; Sevilla, A.; Blanco, S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 240–252. [Google Scholar] [CrossRef] [PubMed]
- David, R.; Burgess, A.; Parker, B.; Li, J.; Pulsford, K.; Sibbritt, T.; Preiss, T.; Searle, I.R. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 2017, 29, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Flores, R. The predominant circular form of avocado sunblotch accumulates in planta as a free RNA adopting a rod-shaped secondary structure unprotected by tightly bound host proteins. J. Gen. Virol. 2017, 98, 1913–1922. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Flores, R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: A “naked” rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017, 14, 1046–1054. [Google Scholar] [CrossRef]
- Rana, A.K.; Ankri, S. Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases. Front Genet. 2016, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Navarro, B.; Flores, F. Origin and evolution of viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA, 2017; Chapter 12; pp. 125–134. ISBN 978-0-12-801498-1. [Google Scholar]
| Name | seq (5′ to 3′) * | Position # |
|---|---|---|
| PSTVd_met_1F_plus | GGGGCGAGGGTGTTTAG | 319–335 |
| PSTVd_met_2R_plus | CACTCCCCACCRTCCTTTTTT | 138–118 |
| PSTVd_met_3F_plus | AAAAAAGGAYGGTGGGGAGTG | 118–138 |
| PSTVd_met_4R_plus | CTAAACACCCTCRCCCC | 335–319 |
| PSTVd_met_5F_minus | GAAGAAAGGAAGGGTGAAAA | 196–177 |
| PSTVd_met_6R_minus | ACCACCCCTCRCCCCCTT | 222–239 |
| ASBVd_met_1F_plus | GTGGTGAAYTTTTATTAAAAAAATTAG | 106–132 |
| ASBVd_met_2R_plus | CCACRACTCCTCCTTCTCTCACAA | 109–86 |
| ASBVd_met_3F_minus | GAGTGAAYTAATTTTTTTAATAAAAGTT | 139–112 |
| ASBVd_met_4R_minus | TCTTCAATCTCTTRATCACTTC | 141–162 |
| HSVd_met_1F_plus | GAGAGGYGTGGAGAGAGGG | 106–125 |
| HSVd_met_2R_plus | CCTCCCTRCCTTATTTTTTCTTT | 56–34 |
| tRNA-Asp_1F | GTCGTTGTAGTATAGTGG | |
| tRNA-Asp_2R | ATCGTTCCCAGGTCAGGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Serio, F.; Torchetti, E.M.; Daròs, J.-A.; Navarro, B. Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level. Viruses 2019, 11, 357. https://doi.org/10.3390/v11040357
Di Serio F, Torchetti EM, Daròs J-A, Navarro B. Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level. Viruses. 2019; 11(4):357. https://doi.org/10.3390/v11040357
Chicago/Turabian StyleDi Serio, Francesco, Enza Maria Torchetti, José-Antonio Daròs, and Beatriz Navarro. 2019. "Reassessment of Viroid RNA Cytosine Methylation Status at the Single Nucleotide Level" Viruses 11, no. 4: 357. https://doi.org/10.3390/v11040357





