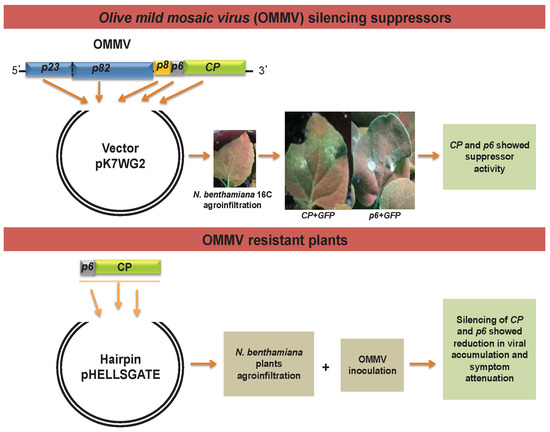
Olive Mild Mosaic Virus Coat Protein and P6 Are Suppressors of RNA Silencing, and Their Silencing Confers Resistance against OMMV
Abstract
:1. Introduction
2. Materials and Methods
2.1. OMMV Silencing Suppressor Genes
2.1.1. Generation of Constructs
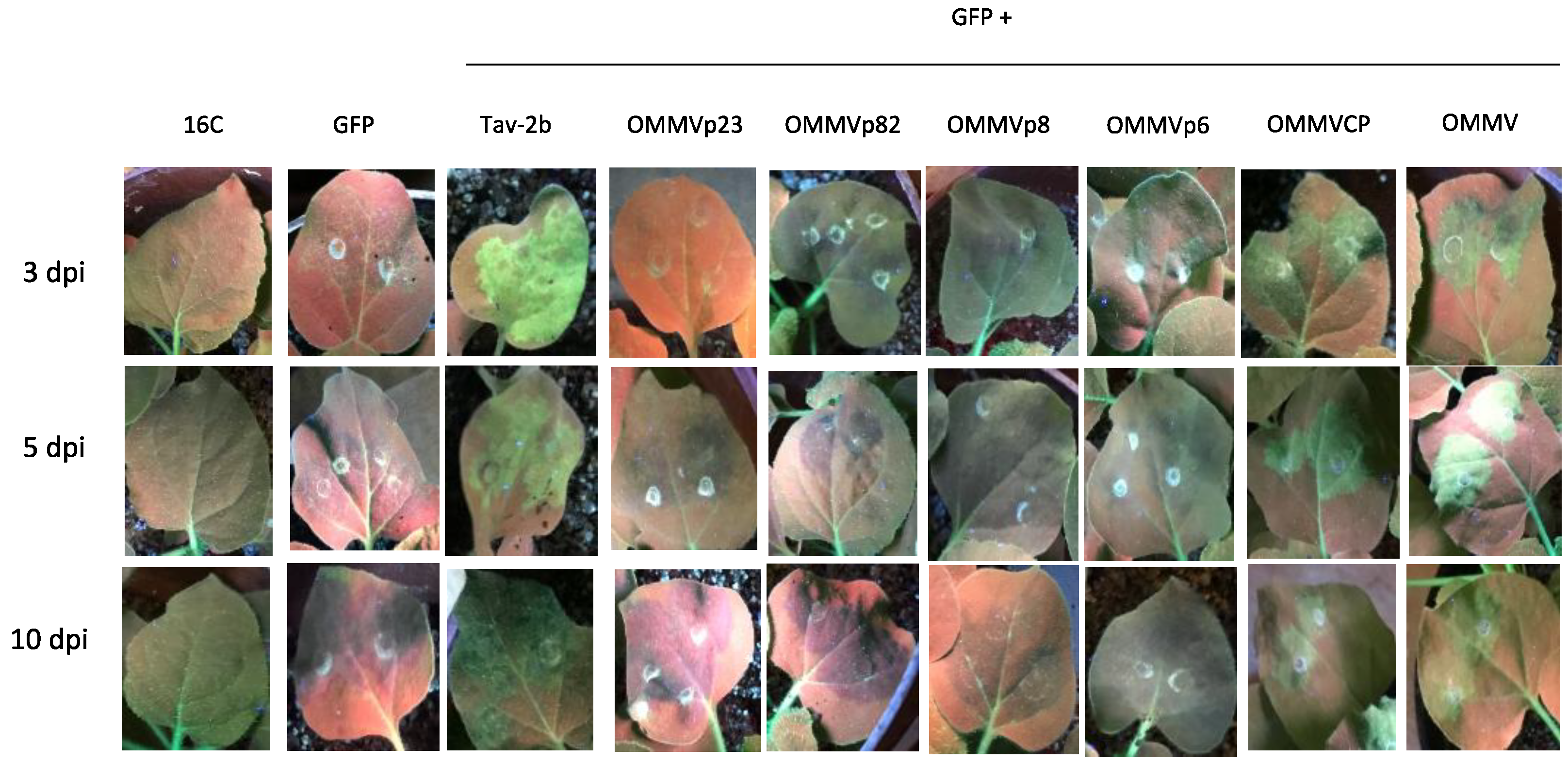
2.1.2. Agrobacterium Co-Infiltration Assays and GFP Imaging
2.1.3. RNA Extraction and Real-Time RT-PCR
2.1.4. Data Analysis
2.2. OMMV Resistance Challenge
3. Results
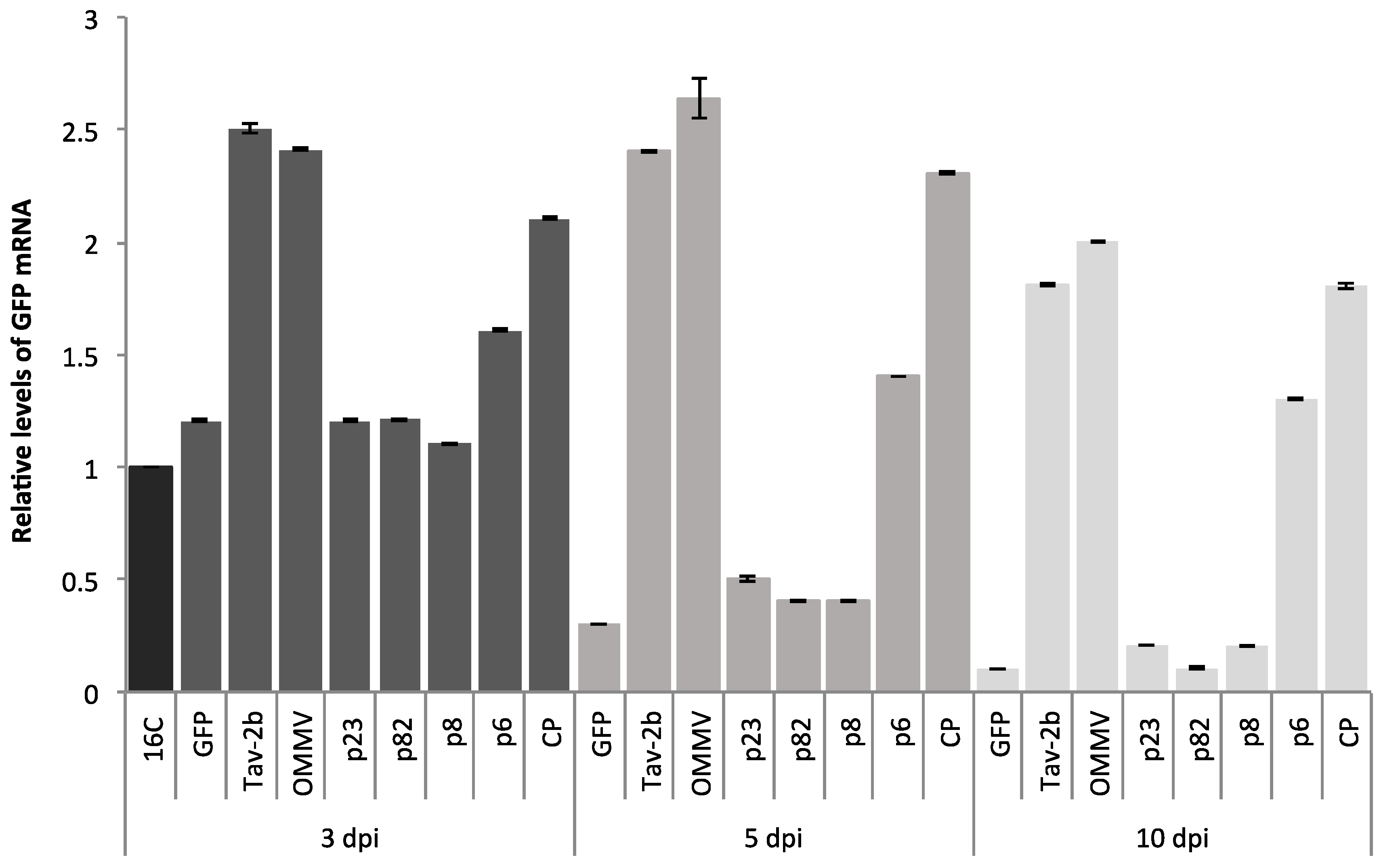
3.1. Determination of OMMV Silencing Suppressors
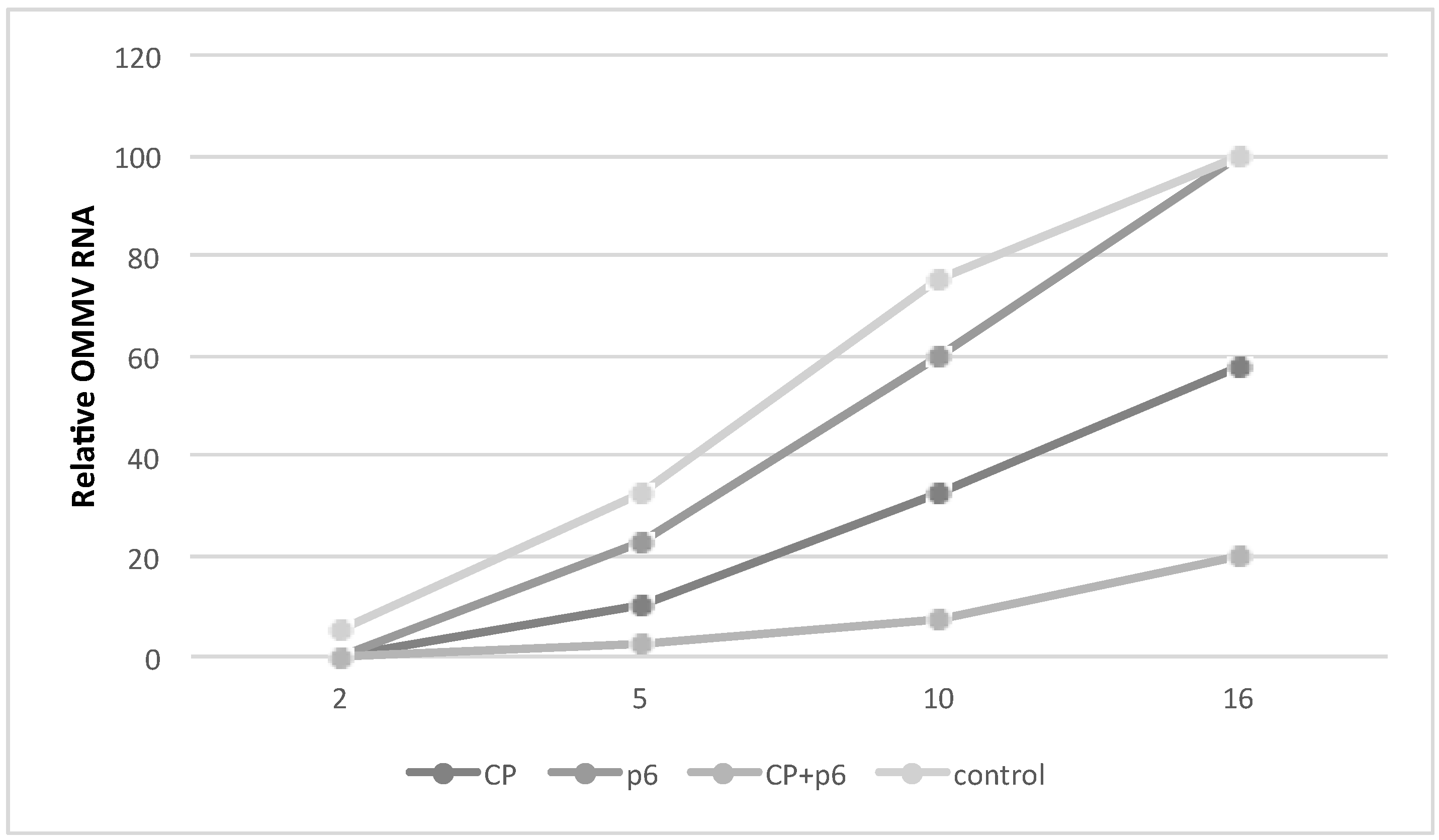
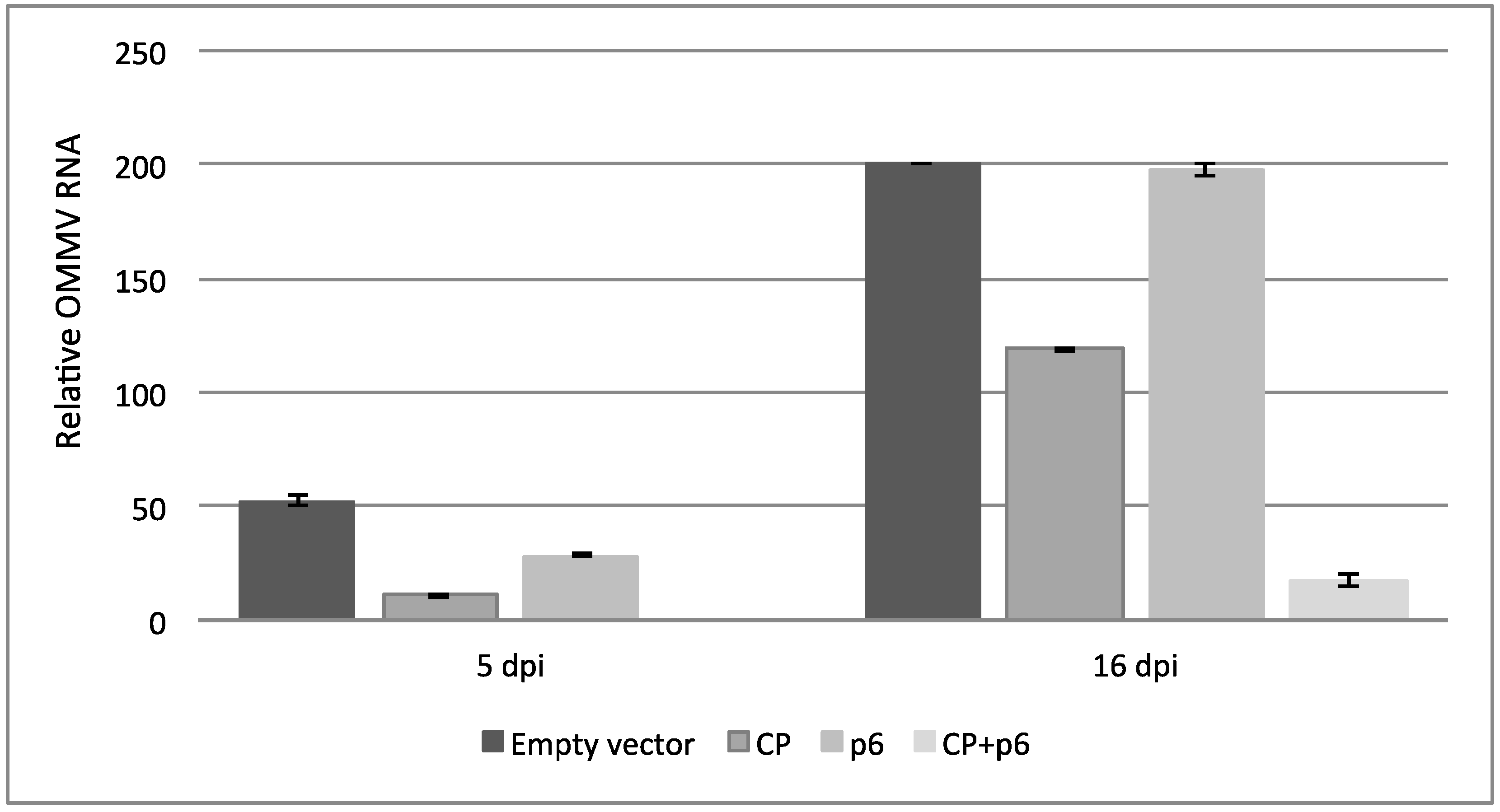
3.2. OMMV Resistance Challenge
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Voinnet, O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001, 17, 449–459. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyán, J. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant argonautes. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyan, J. Viral silencing suppressors: Tools forged to finetune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgyán, J.; Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011, 16, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Samuel, G.H.; Wiley, M.R.; Badawi, A.; Adelman, Z.N.; Myles, K.M. Proceedings of the national academy of sciences of the united states of america. Proc. Natl. Acad. Sci. USA 2016, 113, 13863–13868. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, D.; Molnar, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyan, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, E.J.; Prokhnevsky, A.I.; Gopinath, K.; Dojia, V.V.; Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004, 18, 1179–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; Lopez-Moya, J.J.; Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merai, Z.; Kerenyi, Z.; Molnar, A.; Barta, E.; Valoczi, A.; Bisztray, G.; Havelda, Z.; Burgyan, J.; Silhavy, D. Aureusvirus p14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005, 79, 7217–7226. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ren, T.; Morris, T.J. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 2003, 77, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Simon, A.E. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 2003, 312, 8–13. [Google Scholar] [CrossRef]
- Powers, J.G.; Sit, T.L.; Heinsohn, C.; George, C.G.; Kim, K.-H.; Lommel, S.A. The red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology 2008, 381, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Tsukuda, M.; Mizumoto, H.; Okamoto, K.; Kaido, M.; Mise, K.; Okuno, T. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005, 24, 3147–3157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saponari, M.; Alkowni, R.; Grieco, F.; Pantaleo, V.; Savino, V.; Martelli, G.P.; Driouech, N.; Hassan, M.; Di Terlizzi, B.; Digiaro, M. Detection of olive-infecting viruses in the mediterranean basin. In Proceedings of the Fourth International Symposium on Olive Growing, Valenzano, Italy, 25–30 September 2000; pp. 787–790. [Google Scholar]
- Varanda, C.; Félix, M.R.F.; Leitão, F.; Sismeiro, R.; Clara, M.I.E. Application of reverse transcription—Polymerase chain reaction to screen a collection of clones of olea europaea L. For the presence of necroviruses (tombusviridae). In Proceedings of the 8th Conference of the European Foundation for Plant Pathology & British Society of Plant Pathology Presidential Meeting 2006, Frederiksberg, Denmark, 13–17 August 2006. [Google Scholar]
- El Air, M.; Mahfoudi, N.; Digiaro, M.; Najjar, A.; Elbeaino, T. Detection of olive infecting viruses in tunisia. J. Phytopathol. 2011, 159, 283–286. [Google Scholar] [CrossRef]
- Cardoso, J.; Félix, M.; Clara, M.; Oliveira, S. The complete genome sequence of a new necrovirus isolated from Olea europaea L. Arch. Virol. 2005, 150, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Varanda, C.; Félix, M.; Soares, C.; Oliveira, S.; Clara, M. Specific amino acids of olive mild mosaic virus coat protein are involved on transmission by Olpidium brassicae. J. Gen. Virol. 2011, 92, 2209–2213. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.M.S.; Felix, M.R.; Clara, M.I.E.; Oliveira, S. First characterization of infectious cDNA clones of olive mild mosaic virus. Phytopathol. Mediterr. 2012, 51, 259–265. [Google Scholar]
- Karimi, M.; Inze, D.; Depicker, A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, e193–e195. [Google Scholar] [CrossRef]
- Haseloff, J.; Siemering, K.R.; Prasher, D.C.; Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 1997, 94, 2122–2127. [Google Scholar] [CrossRef] [PubMed]
- Costa, Â.; Marques, N.; Nolasco, G. Citrus tristeza virus p23 may suppress systemic silencing but is not related to the kind of viral syndrome. Physiol. Mol. Plant Pathol. 2014, 87, 69–75. [Google Scholar] [CrossRef]
- Brigneti, G.; Voinnet, O.; Li, W.X.; Ji, L.H.; Ding, S.W.; Baulcombe, D.C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998, 17, 6739–6746. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Warwick, R.M. Changes in Marine Communities: An Approach to Statistical Analysis and Interpretation, 2nd ed.; PRIMER-E Ltd.: Plymouth, UK, 2001. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. Permanova a+ for Primer: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Clarke, K.; Green, R. Statistical design and analysis for a biological effects study. Mar. Ecol. Prog. Ser. 1988, 46, 213–226. [Google Scholar] [CrossRef]
- Zhang, L.; French, R.; Langenberg, W.G. Molecular cloning and sequencing of the coat protein gene of a nebraskan isolate of tobacco necrosis virus. The deduced coat protein sequence has only moderate homology with those of strain A and strain D. Arch. Virol. 1993, 132, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Eamens, A.L.; Waterhouse, P.M. Vectors and methods for hairpin RNA and artificial microRNA-mediated gene silencing in plants. Methods Mol. Biol. 2011, 701, 179–197. [Google Scholar] [PubMed]
- Family-tombusviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 1111–1138.
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.-X.; Falk, B.W.; Dawson, W.O.; Ding, S.-W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.K.; Hein, G.L.; Graybosch, R.A.; Tatineni, S. Octapartite negative-sense RNA genome of high plains wheat mosaic virus encodes two suppressors of RNA silencing. Virology 2018, 518, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, S.; Voinnet, O.; Mette, M.F.; Matzke, M.; Vaucheret, H.; Ding, S.W.; Pruss, G.; Vance, V.B. RNA silencing and the mobile silencing signal. Plant Cell 2002, 14, S289–S301. [Google Scholar] [CrossRef] [PubMed]
- Bag, S.; Mitter, N.; Eid, S.; Pappu, H.R. Complementation between two tospoviruses facilitates the systemic movement of a plant virus silencing suppressor in an otherwise restrictive host. PLoS ONE 2012, 7, e44803. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Ruiz-Ruiz, S.; Soler, N.; Sanchez-Navarro, J.; Fagoaga, C.; Lopez, C.; Navarro, L.; Moreno, P.; Peña, L. Citrus tristeza virus p23: A unique protein mediating key virus-host interactions. Front. Microbiol. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pruss, G.; Ge, X.; Shi, X.M.; Carrington, J.C.; Vance, V.B. Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 1997, 9, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Felix, M.; Varanda, C.; Clara, M. Biology and molecular characterization of necroviruses affecting Olea europaea L.: A review. Eur. J. Plant Pathol. 2012, 133, 247–259. [Google Scholar] [CrossRef]
- Molnar, A.; Havelda, Z.; Dalmay, T.; Szutorisz, H.; Burgyan, J. Complete nucleotide sequence of tobacco necrosis virus strain DH and genes required for RNA replication and virus movement. J. Gen. Virol. 1997, 78, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Lomonossoff, G.P. Pathogen-derived resistance to plant viruses. Ann. Rev. Phytopathol. 1995, 33, 323–343. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.A.; Swaney, S.L.; Parks, T.D.; Wernsman, E.A.; Dougherty, W.G. Transgenic plant virus resistance mediated by untranslatable sense RNAs: Expression, regulation, and fate of nonessential RNAs. Plant Cell 1994, 6, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; Zhu, C.X.; Zhu, X.P.; Guo, X.Q.; Song, Y.Z.; Wen, F.J. A link between PVYN CP gene-mediated virus resistance and transgene arrangement. J. Phytopathol. 2007, 155, 676–682. [Google Scholar] [CrossRef]
- Tenllado, F.; Diaz-Ruiz, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed]
- Simon-Mateo, C.; Garcia, J.A. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A.E. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.A.; Singh, S.P.; Wang, M.B.; Stoutjesdijk, P.A.; Green, A.G.; Waterhouse, P.M. Total silencing by intron-spliced hairpin RNAs. Nature 2000, 407, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Wesley, S.V.; Helliwell, C.A.; Smith, N.; Wang, M.; Rouse, D.T.; Liu, Q.; Gooding, P.S.; Singth, S.P.; Abbott, D.; Stoutjasdijk, P.A.; et al. Construct design for efficient, effective and high throughput gene silencing in plants. Plant J. 2001, 27, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Hily, J.M.; Ravelonandro, M.; Damsteegt, V.; Bassett, C.; Petri, C.; Liu, Z.; Scorza, R. Plum pox virus coat protein gene intron hairpin-RNA (ihpRNA) constructs provide resistance to Plum pox virus in Nicotiana benthamiana and Prunus domestica. J. Am. Soc. Hortic. Sci. 2017, 132, 850–858. [Google Scholar]
- Di Nicola-Negri, E.; Brunetti, A.; Tavazza, M.; Ilardi, V. Hairpin RNA-mediated silencing of Plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgen. Res. 2005, 14, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Sudarshana, M.R.; Ullman, D.E.; Ding, S.W.; Dandekar, A.M.; Falk, B.W. Chimeric cDNA sequences from citrus tristeza virus confer RNA silencing-mediated resistance in transgenic Nicotiana benthamiana plants. Phytopathology 2006, 96, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.L.; Thomas, C.L.; Maule, A.J. De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 1998, 17, 6385–6393. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Fang, Y.Y.; Zhou, B.J.; Zhao, J.H.; Hou, W.N.; Zhu, H.; Ding, S.W.; Guo, H.S. Suppression of arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the cucumber mosaic virus 2b protein. Plant Cell 2012, 24, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Varanda, C.; Cardoso, J.; Félix, M.; Oliveira, S.; Clara, M. Multiplex RT-PCR for detection and identification of three necroviruses that infect olive trees. J. Plant Pathol. 2010, 127, 161–164. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.; Liu, J.; He, H.; Yang, M.; Li, M.; Zhu, X.; Wang, X. A viral suppressor P1/HC-pro increases the GFP gene expression in Agrobacterium-mediated transient assay. Appl. Biochem. Biotechnol. 2009, 158, 243–252. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Primer (5′–3′) | Region Amplified | Location in OMMV Genome | Fragment Length (bp) |
|---|---|---|---|---|
| attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT | - | - | - |
| attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT | |||
| OMMVattB1 | AAAAAGCAGGCTAGTATACATACCAAGTATA | OMMV | 1–19 | 3707 |
| OMMVattB2 | AGAAAGCTGGGTGGGGTCGGGCAAAGGCC | 3667–3683 | ||
| OMMVp23attB1 | AAAAAGCAGGCTAGGATAAAATGGAGCTCAC | p23 | 52–70 | 642 |
| OMMVp23attB2 | AGAAAGCTGGGTCCTATTTGGCCCGGAAGGCC | 650–669 | ||
| OMMVP82attB1 | AAAAAGCAGGCTAGGATAAAATGGAGCTCAC | p82 | 52–70 | 2208 |
| OMMVP82attB2 | AGAAAGCTGGGTATCAGTTTGGTAATCCATTG | 2216–2235 | ||
| OMMVp8attB1 | AAAAAGCAGGCTTTTAATCAATGGATTACCA | p8 | 2210–2228 | 267 |
| OMMVp8attB2 | AGAAAGCTGGGTACACAGCCATAACTCAAAAG | 2433–2452 | ||
| OMMVp6attB1 | AAAAAGCAGGCTCTTTTGAGTTATGGCTGTGT | p6 | 2433–2452 | 208 |
| OMMVp6attB2 | AGAAAGCTGGGTTGTCTATTTTGCGATCG | 2600–2616 | ||
| OMMVCPattB1 | AAAAAGCAGGCTACCAAAACATGCCTAAGAG | CP | 2628–2646 | 842 |
| OMMVCPattB2 | AGAAAGCTGGGTTCAAACGTTAATGGTAGGG | 3427–3445 |
| Gene | Primer Name | Primer (5′–3′) | Reference |
|---|---|---|---|
| Protein phosphatase 2A | PP2ArtFW | GACCCTGATGTTGATGTTCGCT | [30] |
| PP2ArtREV | GAGGGATTTGAAGAGAGATTTC | ||
| F-box protein | FBOXrtFW | GGCACTCACAAACGTCTATTTC | [30] |
| FBOXrtREV | ACCTGGGAGGCATCCTGCTTAT | ||
| GFP | GFP-ER Taq-F | GCCAACACTTGTCACTACTTTCTC | [28] |
| GFP-ER Taq-R | GTAGTTCCCGTCGTCCTTGAAG | ||
| OMMV p23 | OMMVP23rtFW | CGAGTCCGCAAGCAGAAGAAG | This study |
| OMMVP23rtREV | GGGTAGACCAAACTCGGCA | ||
| OMMV p82 | OMMVP82rtFW | TCCAAGACGCCCCGAAAC | This study |
| OMMVP82rtREV | TGGTTACAGGGGAATGACGC | ||
| OMMV p8 | OMMVP8rtFW | GCTCAGAAATCGCAGCAAGG | This study |
| OMMVP8rtREV | TGTCACGGTAATGGTCTGTTCT | ||
| OMMV p6 | OMMVP6rtFW | TGTGTCGCTGCTGTGATACTT | This study |
| OMMVP6rtREV | TTGCAAGGATGAGGATGAGAAT | ||
| OMMV CP | OMMVCPrtFW | TGTCCAGCCACAGCTCTCAT | This study |
| OMMVCPrtREV | TTCGATGAACTCAATCTCATATCGC |
| Pairwise Tests | 3 dpi | 5 dpi | 10 dpi |
|---|---|---|---|
| ORFs | |||
| GFP vs. Tav-2b | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. OMMV | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. p23 | 0.9153 | 0.0001 | 0.0001 |
| GFP vs. p82 | 0.6178 | 0.0001 | 0.3574 |
| GFP vs. p8 | 0.0002 | 0.0001 | 0.0001 |
| GFP vs. p6 | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. CP | 0.0001 | 0.0001 | 0.0001 |
| Tav-2b vs. OMMV | 0.0129 | 0.0518 | 0.0001 |
| Tav-2b vs. p23 | 0.0001 | 0.0001 | 0.0001 |
| Tav-2b vs. p82 | 0.0001 | 0.0001 | 0.0001 |
| Tav-2b vs. p8 | 0.0001 | 0.0001 | 0.0001 |
| Tav-2b vs. p6 | 0.0001 | 0.0001 | 0.0001 |
| Tav-2b vs. CP | 0.0001 | 0.0004 | 0.8368 |
| OMMV vs. p23 | 0.0001 | 0.0001 | 0.0001 |
| OMMV vs. p82 | 0.0001 | 0.0001 | 0.0001 |
| OMMV vs. p8 | 0.0001 | 0.0001 | 0.0001 |
| OMMV vs. p6 | 0.0001 | 0.0002 | 0.0001 |
| OMMV vs. CP | 0.0001 | 0.0134 | 0.0001 |
| p23 vs. p82 | 0.6137 | 0.0016 | 0.6807 |
| p23 vs. p8 | 0.0001 | 0.0017 | 0.6569 |
| p23 vs. p6 | 0.0001 | 0.0001 | 0.0001 |
| p23 vs. CP | 0.0001 | 0.0001 | 0.0001 |
| p82 vs. p8 | 0.0001 | 0.9977 | 0.6799 |
| p82 vs. p6 | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. Tav-2b | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. OMMV | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. p23 | 0.0001 | 0.0001 | 0.0001 |
| GFP vs. p82 | 0.0001 | 0.0001 | 0.0001 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varanda, C.M.; Materatski, P.; Campos, M.D.; Clara, M.I.E.; Nolasco, G.; Félix, M.D.R. Olive Mild Mosaic Virus Coat Protein and P6 Are Suppressors of RNA Silencing, and Their Silencing Confers Resistance against OMMV. Viruses 2018, 10, 416. https://doi.org/10.3390/v10080416
Varanda CM, Materatski P, Campos MD, Clara MIE, Nolasco G, Félix MDR. Olive Mild Mosaic Virus Coat Protein and P6 Are Suppressors of RNA Silencing, and Their Silencing Confers Resistance against OMMV. Viruses. 2018; 10(8):416. https://doi.org/10.3390/v10080416
Chicago/Turabian StyleVaranda, Carla MR, Patrick Materatski, Maria Doroteia Campos, Maria Ivone E. Clara, Gustavo Nolasco, and Maria Do Rosário Félix. 2018. "Olive Mild Mosaic Virus Coat Protein and P6 Are Suppressors of RNA Silencing, and Their Silencing Confers Resistance against OMMV" Viruses 10, no. 8: 416. https://doi.org/10.3390/v10080416