Molecular Characterization of Divergent Closterovirus Isolates Infecting Ribes Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transmission Electron Microscopy
2.2. Genome Assembly and Organization
2.3. In Silico Analyses
3. Results and Discussion
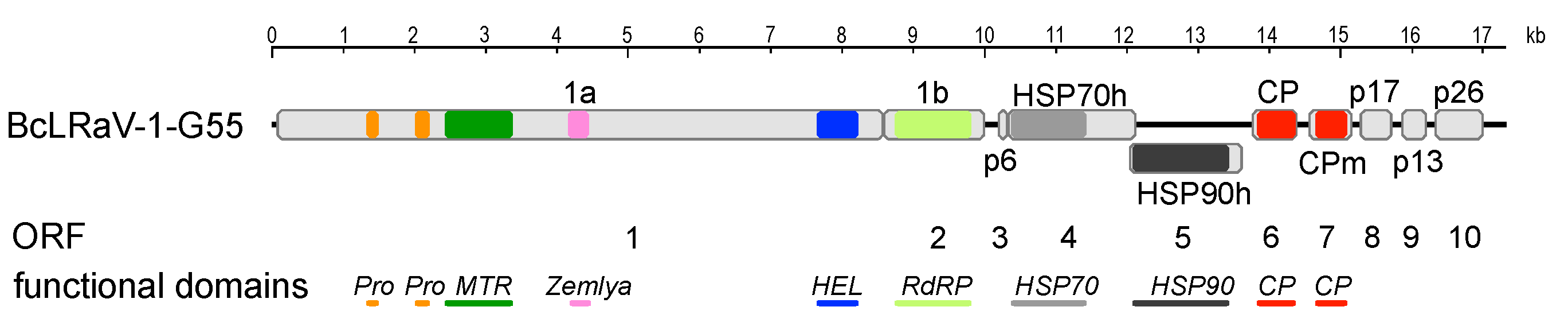
3.1. Sequence and Genome Organization
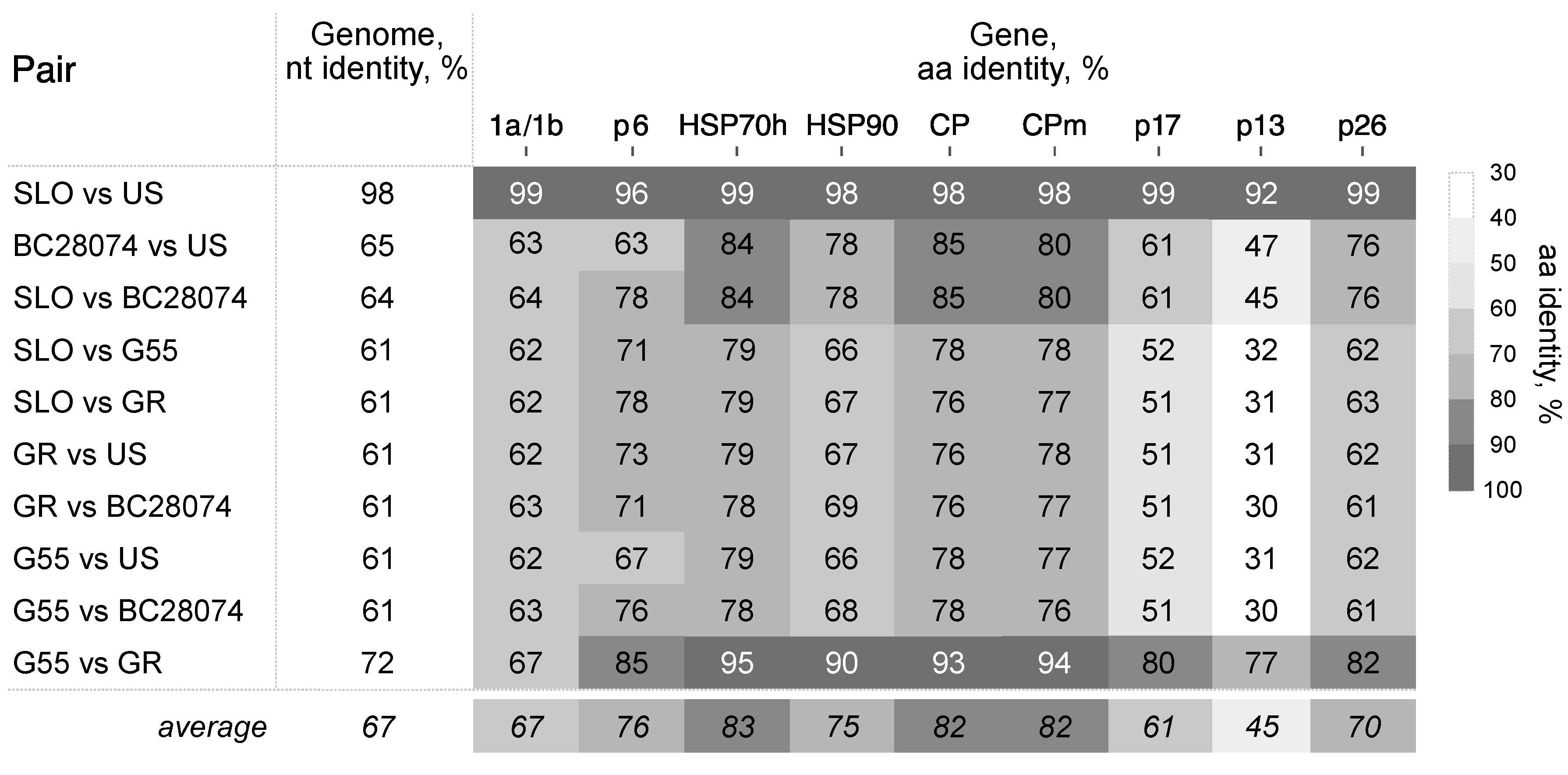
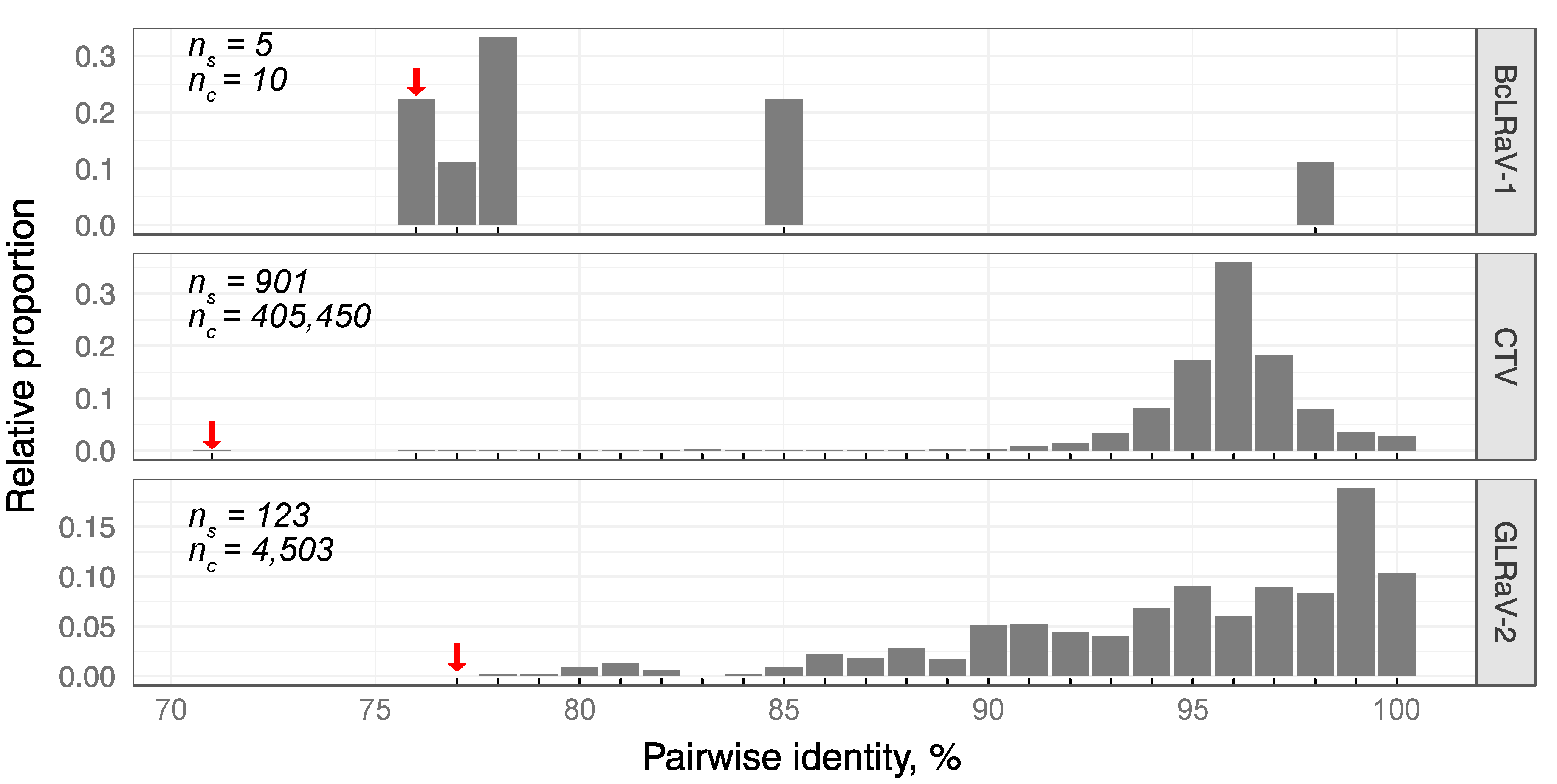
3.2. Divergence of BcLRaV-1
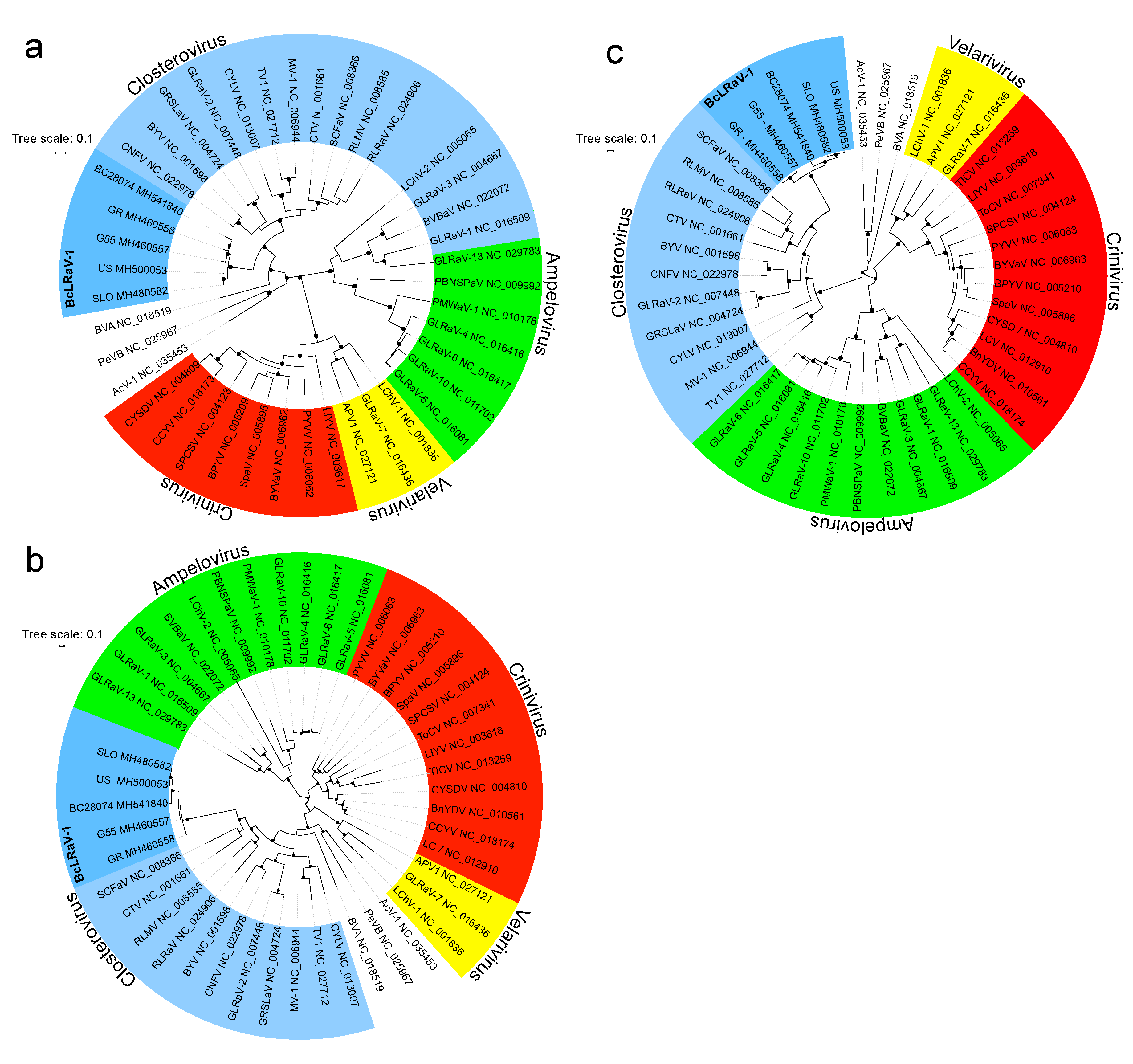
3.3. Phylogenetic Analysis
3.4. Recombination Analysis
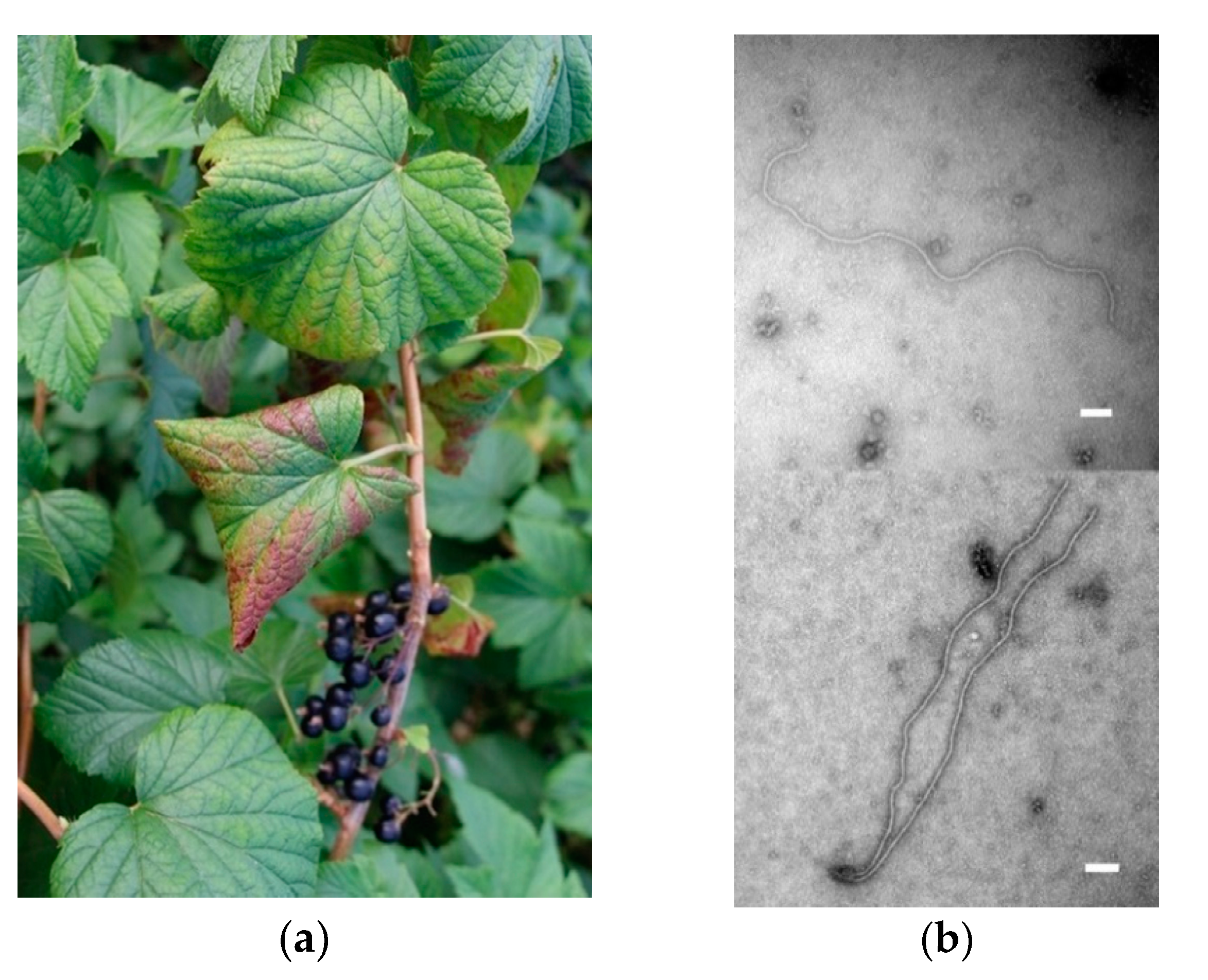
3.5. Transmission Electron Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitchell, C.; Brennan, R.M.; Cross, J.V.; Johnson, S.N. Arthropod pests of currant and gooseberry crops in the U.K.: Their biology, management and future prospects. Agric. For. Entomol. 2011, 13, 221–237. [Google Scholar] [CrossRef]
- Geils, B.W.; Hummer, K.E.; Hunt, R.S. White pines, Ribes, and blister rust: A review and synthesis. For. Pathol. 2010, 40, 147–185. [Google Scholar] [CrossRef]
- Adams, A.N.; Thresh, J.M. Reversion of black currant. In Virus Disease of Small Fruits; Converse, R.H., Ed.; United States Department of Agriculture: Washington, DC, USA, 1987; pp. 133–136. [Google Scholar]
- Jones, A.T. Black currant reversion disease—The probable causal agent, eriophyid mite vectors, epidemiology and prospects for control. Virus Res. 2000, 71, 71–84. [Google Scholar] [CrossRef]
- Jones, A.T.; McGavin, W.J.; Geering, A.D.W.; Lockhart, B.E.L. A new badnavirus in Ribes species, its detection by PCR, and its close association with gooseberry vein banding disease. Plant Dis. 2001, 85, 417–422. [Google Scholar] [CrossRef]
- Besse, S.; Gugerli, P.; Ramel, M.E.; Balmelli, C. Characterisation of mixed virus infections in Ribes species in Switzerland. In Proceedings of the 21st International Conference on Virus and other Graft Transmissible Diseases of Fruit Crops, Neustadt, Germany, 5–10 July 2009; pp. 214–219. [Google Scholar]
- Ho, T.; Tzanetakis, I.E. Development of a virus detection and discovery pipeline using next generation sequencing. Virology 2014, 471–473, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Petrzik, K.; Koloniuk, I.; Přibylová, J.; Špak, J. Complete genome sequence of currant latent virus (genus Cheravirus, family Secoviridae). Arch. Virol. 2015, 161, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Petrzik, K.; Přibylová, J.; Koloniuk, I.; Špak, J. Molecular characterization of a novel capillovirus from red currant. Arch. Virol. 2016, 161, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Phelan, J. Complete genome sequence of a strain of Actinidia virus X detected in Ribes nigrum cv. Baldwin showing unusual symptoms. Arch. Virol. 2016, 161, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.P.; Yang, T.; Liu, H.W.; Postman, J.; Li, R. Molecular characterization of a novel rhabdovirus infecting blackcurrant identified by high-throughput sequencing. Arch. Virol. 2018, 162, 2493–2494. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.M.; Jones, A.T. Rhabdovirus-like and closterovirus-like particles in ultrathin sections of Ribes species with symptoms of blackcurrant reversion and gooseberry veinbanding diseases. Ann. Appl. Biol. 1997, 130, 77–89. [Google Scholar] [CrossRef]
- Ho, T.; Postman, J.D.; Tzanetakis, I.E. Discovery, characterization and detection of five new virus species in Ribes. In Proceedings of the 23rd International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Morioka, Japan, 8–12 June 2015; PSJ Plant Virus Disease Workshop Report No. 12 (Special Edition). Phytopathological Society of Japan: Tokyo, Japan, 2016; pp. 8–10. [Google Scholar]
- Rubio, L.; Guerri, J.; Moreno, P. Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front. Microbiol. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P.; Agranovsky, A.A.; Bar-Joseph, M.; Boscia, D.; Candresse, T.; Coutts, R.H.A.; Dolja, V.V.; Hu, J.S.; Jelkmann, W.; Karasev, A.V.; et al. Family Closteroviridae. In Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: New York, NY, USA, 2012; pp. 987–1001. [Google Scholar]
- Gugerli, P.; Ramel, M.E. Production of monoclonal antibodies for the serological identification and reliable detection of Apple stem pitting and Pear yellow vein viruses in apple and pear. Acta Hortic. 2004, 59–69. [Google Scholar] [CrossRef]
- Koloniuk, I.; Fránová, J.; Sarkisova, T.; Přibylová, J.; Lenz, O.; Petrzik, K.; Špak, J. Identification and molecular characterization of a novel varicosa-like virus from red clover. Arch. Virol. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Gugerli, P. Isopycnic centrifugation of plant-viruses in nycodenz density gradients. J. Virol. Methods 1984, 9, 249–258. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.W.; Peremyslov, V.V.; Mushegian, A.R.; Dawson, W.O.; Dolja, V.V. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 2001, 75, 12153–12160. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-P.; Peremyslov, V.V.; Medina, V.; Dolja, V.V. Tandem leader proteases of Grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology 2009, 383, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Karlin, D.G.; Makhotenko, A.V.; Khromov, A.V.; Erokhina, T.N.; Solovyev, A.G.; Morozov, S.Y.; Agranovsky, A.A. A conserved region in the Closterovirus 1a polyprotein drives extensive remodeling of endoplasmic reticulum membranes and induces motile globules in Nicotiana benthamiana cells. Virology 2016, 502, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef] [PubMed]
- Erokhina, T.N.; Zinovkin, R.A.; Vitushkina, M.V.; Jelkmann, W.; Agranovsky, A.A. Detection of beet yellows closterovirus methyltransferase-like and helicase-like proteins in vivo using monoclonal antibodies. J. Gen. Virol. 2000, 81, 597–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peremyslov, V.V.; Pan, Y.W.; Dolja, V.V. Movement protein of a closterovirus Is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 2004, 78, 3704–3709. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.-X.; Falk, B.W.; Dawson, W.O.; Ding, S.-W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynard, J.-S.; Schneeberger, P.H.H.; Frey, J.E.; Schaerer, S. Biological, serological, and molecular characterization of a highly divergent strain of Grapevine leafroll-associated virus 4 causing grapevine leafroll disease. Phytopathology 2015, 105, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Štrukelj, M.; Pleško, I.M.; Urek, G. Molecular characterization of a grapevine leafroll-associated virus 4 from Slovenian vineyards. Acta Virol. 2016, 60, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Joseph, M.; Dawson, W.O. Citrus Tristeza Virus. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 160–165. [Google Scholar]
- Martín, S.; Sambade, A.; Rubio, L.; Vives, M.C.; Moya, P.; Guerri, J.; Elena, S.F.; Moreno, P. Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J. Gen. Virol. 2009, 90, 1527–1538. [Google Scholar] [CrossRef] [PubMed]

| BcLRaV-1 Isolate/GenBank Accession Number | Plant | Origin | Symptoms | Sequencing | |
|---|---|---|---|---|---|
| Input | HTS Output | ||||
| G55/MH460557 | Red currant, Gabreta 55 | Czech Republic | Asymptomatic | Total RNA: mRNA enriched | 20 millions, 100 bp reads |
| GR/MH460558 | Red currant, Gondouin Rouge | Czech Republic | Asymptomatic | ||
| SLO/MH480582 | Black currant, unknown cultivar | Slovenia | Asymptomatic | Total RNA: Ribo-depleted | 10 millions, 2 × 150 nt reads |
| BC28074/MH541840 | Black currant, 28074 | Switzerland | Leafroll [6] | Viral-associated nucleic acid | 50 millions, 2 × 75 nt reads |
| US/MH500053 | Black currant, NCGR PI 556169 | USA | Yellow line patterns [13] | Enriched double-stranded RNA | 76,214 reads |
| Isolate | Genome Length (nt) | Genome Elements | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′-UTR (nt) | 3′-UTR (nt) | Length of ORFs for Predicted Proteins (nt)/ Molecular Mass of Encoded Proteins (kDa) | ||||||||||
| 1a/1b | p6 | HSP70h | HSP90 | CP | CPm | p17 | p13 | p26 | ||||
| G55 | 17,313 | 97 | 275 | 9939/371.9 | 147/5.7 | 1797/65.6 | 1593/61 | 636/23.6 | 606/22.3 | 456/16.8 | 348/13.1 | 690/25.6 |
| GR | 17,161 | 99 | 309 | 9906/370.5 | 147/5.7 | 1797/65.5 | 1590/60.9 | 636/23.6 | 606/22.2 | 441/16.3 | 348/13.2 | 690/25.8 |
| SLO | 16,894 | 1 * | 260 * | 9942/372.7 | 150/5.6 | 1797/65.7 | 1593/60.6 | 636/23.4 | 606/22.5 | 444/16.5 | 372/14.3 | 687/25.5 |
| BC28074 | 17,141 | 102 | 290 | 9942/370.3 | 150/5.7 | 1797/65.9 | 1593/60.4 | 636/23.5 | 714/26.7 | 444/16.4 | 399/15 | 687/25.6 |
| US | 16,996 | 99 | 264 | 9942/372.7 | 150/5.6 | 1797/65.7 | 1593/60.7 | 636/23.4 | 606/22.5 | 444/16.6 | 372/14.2 | 687/25.5 |
| Predicted Recombinant Isolate | Position of the Recombinant Part (Predicted Breakpoints) | Putative Parental Isolates 1 | Detection Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Major Parent | Minor Parent | RDP | GENECONV | Bootscan | Maxchi | Chimaera | SiSscan | PhylPro | LARD | 3Seq | ||
| G55 | 45–9957 | Unknown (US) | GR | 1.1 × 10−52 | NS 2 | 5.1 × 10−40 | 2.2 × 10−15 | 4.6 × 10-33 | 2.3 × 10-84 | NS 2 | NS 2 | 1.6 × 10−5 |
| US | 7701–10,345 | BC28074 | GR | 2.7 × 10−5 | NS 2 | 2.3 × 10−2 | 1.5 × 10−4 | 7.5 × 10−6 | 5.8 × 10−4 | NS 2 | 4.3 × 10-51 | 2.2 × 10−5 |
| US | 4514–4729 | GR | G55 | 1.0 × 10−3 | 2.5 × 10−2 | 7.2 × 10−3 | NS 2 | 4.3 × 10−2 | 8.5 × 10−9 | NS 2 | NS 2 | 4.1 × 10−2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koloniuk, I.; Thekke-Veetil, T.; Reynard, J.-S.; Mavrič Pleško, I.; Přibylová, J.; Brodard, J.; Kellenberger, I.; Sarkisova, T.; Špak, J.; Lamovšek, J.; et al. Molecular Characterization of Divergent Closterovirus Isolates Infecting Ribes Species. Viruses 2018, 10, 369. https://doi.org/10.3390/v10070369
Koloniuk I, Thekke-Veetil T, Reynard J-S, Mavrič Pleško I, Přibylová J, Brodard J, Kellenberger I, Sarkisova T, Špak J, Lamovšek J, et al. Molecular Characterization of Divergent Closterovirus Isolates Infecting Ribes Species. Viruses. 2018; 10(7):369. https://doi.org/10.3390/v10070369
Chicago/Turabian StyleKoloniuk, Igor, Thanuja Thekke-Veetil, Jean-Sébastien Reynard, Irena Mavrič Pleško, Jaroslava Přibylová, Justine Brodard, Isabelle Kellenberger, Tatiana Sarkisova, Josef Špak, Janja Lamovšek, and et al. 2018. "Molecular Characterization of Divergent Closterovirus Isolates Infecting Ribes Species" Viruses 10, no. 7: 369. https://doi.org/10.3390/v10070369





