Gp120 V5 Is Targeted by the First Wave of Sequential Neutralizing Antibodies in SHIVSF162P3N-Infected Rhesus Macaques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells, Plasmids and Viruses
2.3. SHIV env Single Genome Amplification and Cloning
2.4. Viral Neutralization Assay
2.5. Site-Directed Mutagenesis
2.6. Statistical Analysis
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Comparisons between SHIV-Infected Macaques with and without HIV-1 Neutralization Breadth
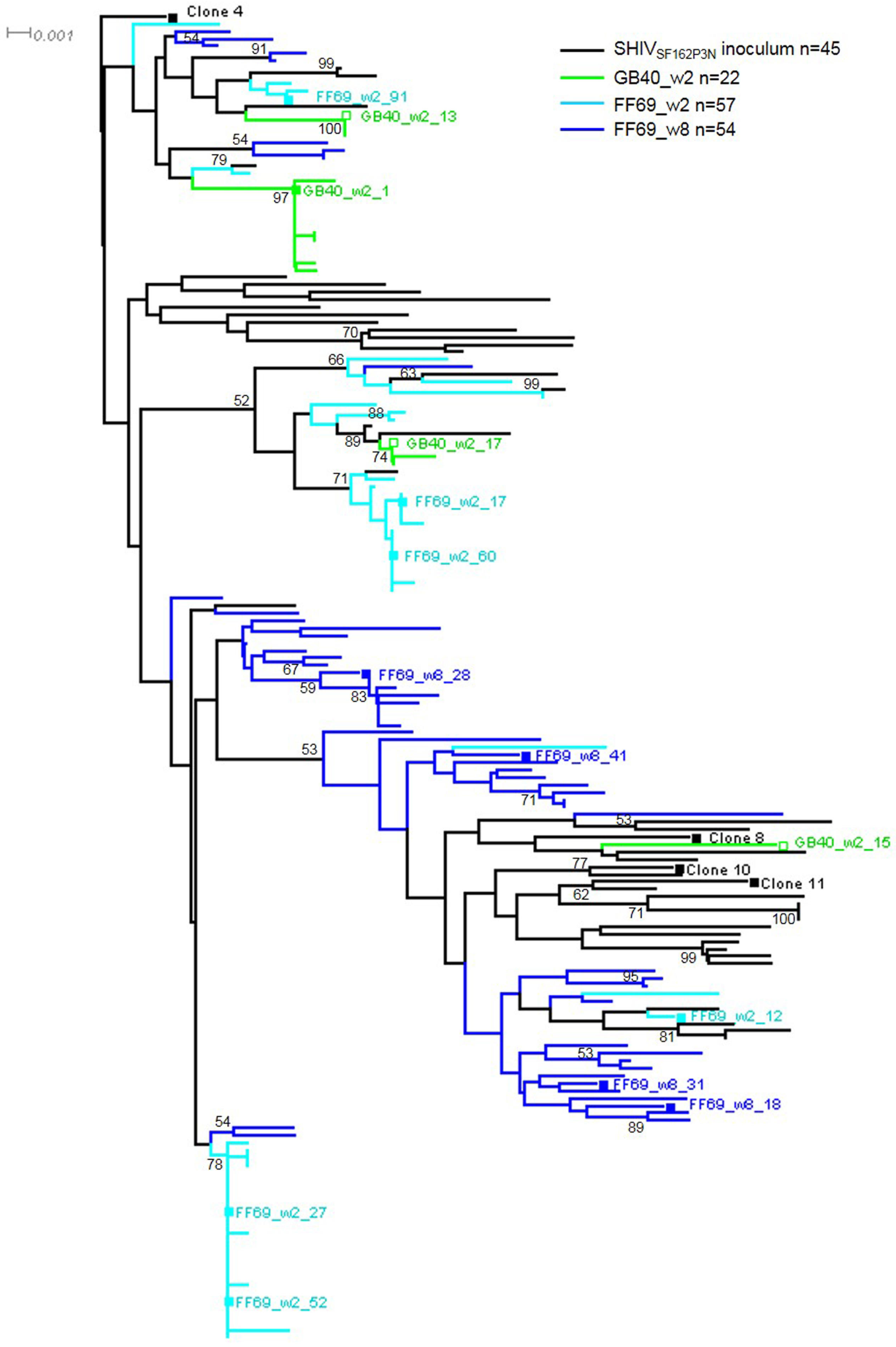
3.2. Transmitted/Founder env Sequences in SHIV-Infected Macaques
3.3. Sequential Neutralizing Antibody Development and Neutralization Sensitivity of the T/F Env Strains
3.4. Targets of Wave 1 Neutralizing Antibodies in SHIV-Infected Macaques
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kwong, P.D.; Mascola, J.R.; Nabel, G.J. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: The end of the beginning. Nat. Rev. Immunol. 2013, 13, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Mascola, J.R.; Haynes, B.F. HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol. Rev. 2013, 254, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R.; Mascola, J.R. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 2015, 16, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Mikell, I.; Sather, D.N.; Kalams, S.A.; Altfeld, M.; Alter, G.; Stamatatos, L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011, 7, e1001251. [Google Scholar] [CrossRef]
- Hraber, P.; Seaman, M.S.; Bailer, R.T.; Mascola, J.R.; Montefiori, D.C.; Korber, B.T. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 2014, 28, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Lu, H.; Markowitz, M.; Cheng-Mayer, C.; Wu, X. Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J. Virol. 2016, 90, 4017–4031. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.X.; Lynch, R.; Zhou, T.; Gao, F.; Alam, S.M.; Boyd, S.D.; Fire, A.Z.; Roskin, K.M.; Schramm, C.A.; Zhang, Z.; et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 2013, 496, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Doria-Rose, N.A.; Schramm, C.A.; Gorman, J.; Moore, P.L.; Bhiman, J.N.; DeKosky, B.J.; Ernandes, M.J.; Georgiev, I.S.; Kim, H.J.; Pancera, M.; et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 2014, 509, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Bonsignori, M.; Liao, H.X.; Kumar, A.; Xia, S.M.; Lu, X.; Cai, F.; Hwang, K.K.; Song, H.; Zhou, T.; et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 2014, 158, 481–491. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, D.T.; Choi, N.M.; Briney, B.; Garces, F.; Ver, L.S.; Landais, E.; Murrell, B.; Wrin, T.; Kilembe, W.; Liang, C.H.; et al. Early antibody lineage diversification and independent LIMB maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 2016, 44, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Bonsignori, M.; Kreider, E.F.; Fera, D.; Meyerhoff, R.R.; Bradley, T.; Wiehe, K.; Alam, S.M.; Aussedat, B.; Walkowicz, W.E.; Hwang, K.K.; et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.L.; Gray, E.S.; Wibmer, C.K.; Bhiman, J.N.; Nonyane, M.; Sheward, D.J.; Hermanus, T.; Bajimaya, S.; Tumba, N.L.; Abrahams, M.R.; et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 2012, 18, 1688–1692. [Google Scholar] [CrossRef] [PubMed]
- Bhiman, J.N.; Anthony, C.; Doria-Rose, N.A.; Karimanzira, O.; Schramm, C.A.; Khoza, T.; Kitchin, D.; Botha, G.; Gorman, J.; Garrett, N.J.; et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 2015, 21, 1332–1336. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Kilby, J.M.; Saag, M.S.; et al. Antibody neutralization and escape by HIV-1. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Richman, D.D.; Wrin, T.; Little, S.J.; Petropoulos, C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 2003, 100, 4144–4149. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Li, B.; Lynch, R.M.; Haaland, R.E.; Murphy, M.K.; Mulenga, J.; Allen, S.A.; Pinter, A.; Shaw, G.M.; Hunter, E.; et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009, 5, e1000594. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.L.; Ranchobe, N.; Lambson, B.E.; Gray, E.S.; Cave, E.; Abrahams, M.R.; Bandawe, G.; Mlisana, K.; Abdool Karim, S.S.; Williamson, C.; et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009, 5, e1000598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, M.K.; Yue, L.; Pan, R.; Boliar, S.; Sethi, A.; Tian, J.; Pfafferot, K.; Karita, E.; Allen, S.A.; Cormier, E.; et al. Viral escape from neutralizing antibodies in early subtype a HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog. 2013, 9, e1003173. [Google Scholar] [CrossRef] [PubMed]
- Lynch, R.M.; Rong, R.; Boliar, S.; Sethi, A.; Li, B.; Mulenga, J.; Allen, S.; Robinson, J.E.; Gnanakaran, S.; Derdeyn, C.A. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J. Virol. 2011, 85, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.S.; Moody, M.A.; Wibmer, C.K.; Chen, X.; Marshall, D.; Amos, J.; Moore, P.L.; Foulger, A.; Yu, J.S.; Lambson, B.; et al. Isolation of a monoclonal antibody that targets the α-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J. Virol. 2011, 85, 7719–7729. [Google Scholar] [CrossRef] [PubMed]
- Lifson, J.D.; Haigwood, N.L. Lessons in nonhuman primate models for aids vaccine research: From minefields to milestones. Cold Spring Harbor Perspect. Med. 2012, 2, a007310. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Kong, R.; Ding, W.; Lee, F.H.; Parker, Z.; Kim, E.; Learn, G.H.; Hahn, P.; Policicchio, B.; et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA 2016, 113, E3413–E3422. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.M.; Sok, D.; Nishimura, Y.; Donau, O.; Sadjadpour, R.; Gautam, R.; Shingai, M.; Pejchal, R.; Ramos, A.; Simek, M.D.; et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc. Natl. Acad. Sci. USA 2011, 108, 20125–20129. [Google Scholar] [CrossRef] [PubMed]
- Shingai, M.; Donau, O.K.; Schmidt, S.D.; Gautam, R.; Plishka, R.J.; Buckler-White, A.; Sadjadpour, R.; Lee, W.R.; LaBranche, C.C.; Montefiori, D.C.; et al. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc. Natl. Acad. Sci. USA 2012, 109, 19769–19774. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Wang, W.; Wang, C.; Gu, T.; Guo, R.; Yu, B.; Kong, W.; Qin, C.; Giorgi, E.E.; Chen, Z.; et al. Development of broad neutralization activity in simian/human immunodeficiency virus-infected rhesus macaques after long-term infection. AIDS 2018, 32, 555–563. [Google Scholar] [PubMed]
- Ho, S.H.; Tasca, S.; Shek, L.; Li, A.; Gettie, A.; Blanchard, J.; Boden, D.; Cheng-Mayer, C. Coreceptor switch in r5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 2007, 81, 8621–8633. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Mumbauer, A.; Zhuang, K.; Harbison, C.; Knight, H.; Westmoreland, S.; Gettie, A.; Blanchard, J.; Cheng-Mayer, C. Mucosal transmissibility, disease induction and coreceptor switching of R5 SHIVSF162P3N molecular clones in rhesus macaques. Retrovirology 2013, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, W.; Tasca, S.; Zhuang, K.; Gettie, A.; Blanchard, J.; Cheng-Mayer, C. Different tempo and anatomic location of dual-tropic and X4 virus emergence in a model of R5 simian-human immunodeficiency virus infection. J. Virol. 2010, 84, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Shakirzyanova, M.; Tsai, L.; Ren, W.; Gettie, A.; Blanchard, J.; Cheng-Mayer, C. Pathogenic consequences of vaginal infection with CCR5-tropic simian-human immunodeficiency virus SHIVSF162P3N. J. Virol. 2012, 86, 9432–9442. [Google Scholar] [CrossRef] [PubMed]
- Mumbauer, A.; Gettie, A.; Blanchard, J.; Cheng-Mayer, C. Efficient mucosal transmissibility but limited pathogenicity of R5 SHIV SF162P3N in chinese-origin rhesus macaques. J. Acquir. Immune Defic. Syndr. 2013, 62, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Decker, J.M.; Liu, H.; Zhang, Z.; Arani, R.B.; Kilby, J.M.; Saag, M.S.; Wu, X.; Shaw, G.M.; Kappes, J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002, 46, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Gonzalez, J.F.; Bailes, E.; Pham, K.T.; Salazar, M.G.; Guffey, M.B.; Keele, B.F.; Derdeyn, C.A.; Farmer, P.; Hunter, E.; Allen, S.; et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008, 82, 3952–3970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Wang, C.; O’Dell, S.; Li, Y.; Keele, B.F.; Yang, Z.; Imamichi, H.; Doria-Rose, N.; Hoxie, J.A.; Connors, M.; et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J. Virol. 2012, 86, 5844–5856. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, K.; Leda, A.R.; Tsai, L.; Knight, H.; Harbison, C.; Gettie, A.; Blanchard, J.; Westmoreland, S.; Cheng-Mayer, C. Emergence of CD4 independence envelopes and astrocyte infection in R5 simian-human immunodeficiency virus model of encephalitis. J. Virol. 2014, 88, 8407–8420. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.S.; Janes, H.; Hawkins, N.; Grandpre, L.E.; Devoy, C.; Giri, A.; Coffey, R.T.; Harris, L.; Wood, B.; Daniels, M.G.; et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010, 84, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, T.; O’Dell, S.; Wyatt, R.T.; Kwong, P.D.; Mascola, J.R. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 2009, 83, 10892–10907. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Jones, G.B.; Soto, C.; Lemmin, T.; Chuang, G.Y.; Druz, A.; Kong, R.; Thomas, P.V.; Wagh, K.; Zhou, T.; Behrens, A.J.; et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 2016, 165, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.S.; Madiga, M.C.; Hermanus, T.; Moore, P.L.; Wibmer, C.K.; Tumba, N.L.; Werner, L.; Mlisana, K.; Sibeko, S.; Williamson, C.; et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 2011, 85, 4828–4840. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.M.; Hunter, E. HIV transmission. Cold Spring Harbor Perspect. Med. 2012, 2, a006965. [Google Scholar] [CrossRef] [PubMed]
- Ronen, K.; Sharma, A.; Overbaugh, J. HIV transmission biology: Translation for HIV prevention. AIDS 2015, 29, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, S.M.; Selhorst, P.; Arien, K.K.; Dorfman, J.R. The HIV-1 transmission bottleneck. Retrovirology 2017, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Jardine, J.; Julien, J.P.; Menis, S.; Ota, T.; Kalyuzhniy, O.; McGuire, A.; Sok, D.; Huang, P.S.; MacPherson, S.; Jones, M.; et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 2013, 340, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Jardine, J.G.; Kulp, D.W.; Havenar-Daughton, C.; Sarkar, A.; Briney, B.; Sok, D.; Sesterhenn, F.; Ereno-Orbea, J.; Kalyuzhniy, O.; Deresa, I.; et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016, 351, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Escolano, A.; Steichen, J.M.; Dosenovic, P.; Kulp, D.W.; Golijanin, J.; Sok, D.; Freund, N.T.; Gitlin, A.D.; Oliveira, T.; Araki, T.; et al. Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 2016, 166, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Briney, B.; Sok, D.; Jardine, J.G.; Kulp, D.W.; Skog, P.; Menis, S.; Jacak, R.; Kalyuzhniy, O.; de Val, N.; Sesterhenn, F.; et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 2016, 166, 1459–1470. [Google Scholar] [CrossRef] [PubMed]



| Macaque ID | Time Point | Sample Type | CD4 Count Cells/μL | Plasma vRNA Copies/mL | No. of Env SGA | Mean ± SD Distance (%) | Maximum Distance (%) | No. of Env Clones | Env Clones | Wave 1 Neutralization |
|---|---|---|---|---|---|---|---|---|---|---|
| GB40 | 2 wpi | Plasma | 1904 | 4,276,200 | 22 | 0.74 ± 0.63 | 2.12 | 4 | w2_1 | Sensitive |
| w2_13,15,17 | Resistant | |||||||||
| FF69 | 2 wpi | Plasma | 269 | 37,524,600 | 57 | 1.01 ± 0.62 | 2.17 | 6 | w2_12,17,27,52,60,91 | Sensitive |
| FF69 | 8 wpi | Plasma | 348 | 775,130 | 54 | 1.15 ± 0.36 | 2.33 | 4 | w8_18,28,31,41 | Sensitive |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, M.; Lu, H.; Kong, X.-P.; Cheng-Mayer, C.; Wu, X. Gp120 V5 Is Targeted by the First Wave of Sequential Neutralizing Antibodies in SHIVSF162P3N-Infected Rhesus Macaques. Viruses 2018, 10, 262. https://doi.org/10.3390/v10050262
Jia M, Lu H, Kong X-P, Cheng-Mayer C, Wu X. Gp120 V5 Is Targeted by the First Wave of Sequential Neutralizing Antibodies in SHIVSF162P3N-Infected Rhesus Macaques. Viruses. 2018; 10(5):262. https://doi.org/10.3390/v10050262
Chicago/Turabian StyleJia, Manxue, Hong Lu, Xiang-Peng Kong, Cecilia Cheng-Mayer, and Xueling Wu. 2018. "Gp120 V5 Is Targeted by the First Wave of Sequential Neutralizing Antibodies in SHIVSF162P3N-Infected Rhesus Macaques" Viruses 10, no. 5: 262. https://doi.org/10.3390/v10050262





