An Immunodominant Region of the Envelope Glycoprotein of Small Ruminant Lentiviruses May Function as Decoy Antigen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Synthetic Peptides
2.3. Antibody Affinity Purification
2.4. Peptide-ELISA and Anti-SU5 Titer
2.5. Antibody Avidity Measurements
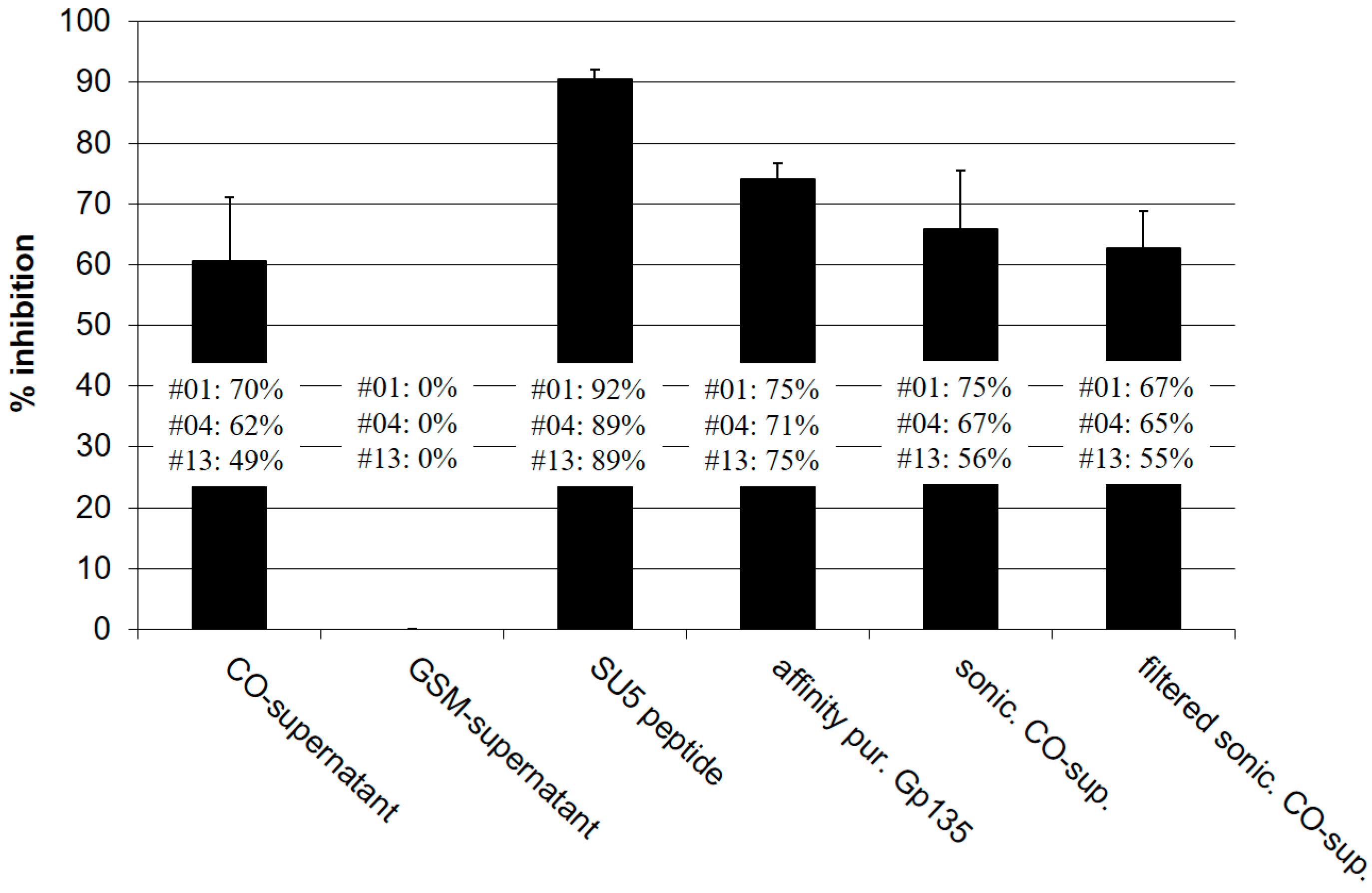
2.6. Antibody Binding Inhibition Assays
2.7. Virus Neutralization
2.8. Sequence Alignments
3. Results
3.1. Antibody Titer and Avidity
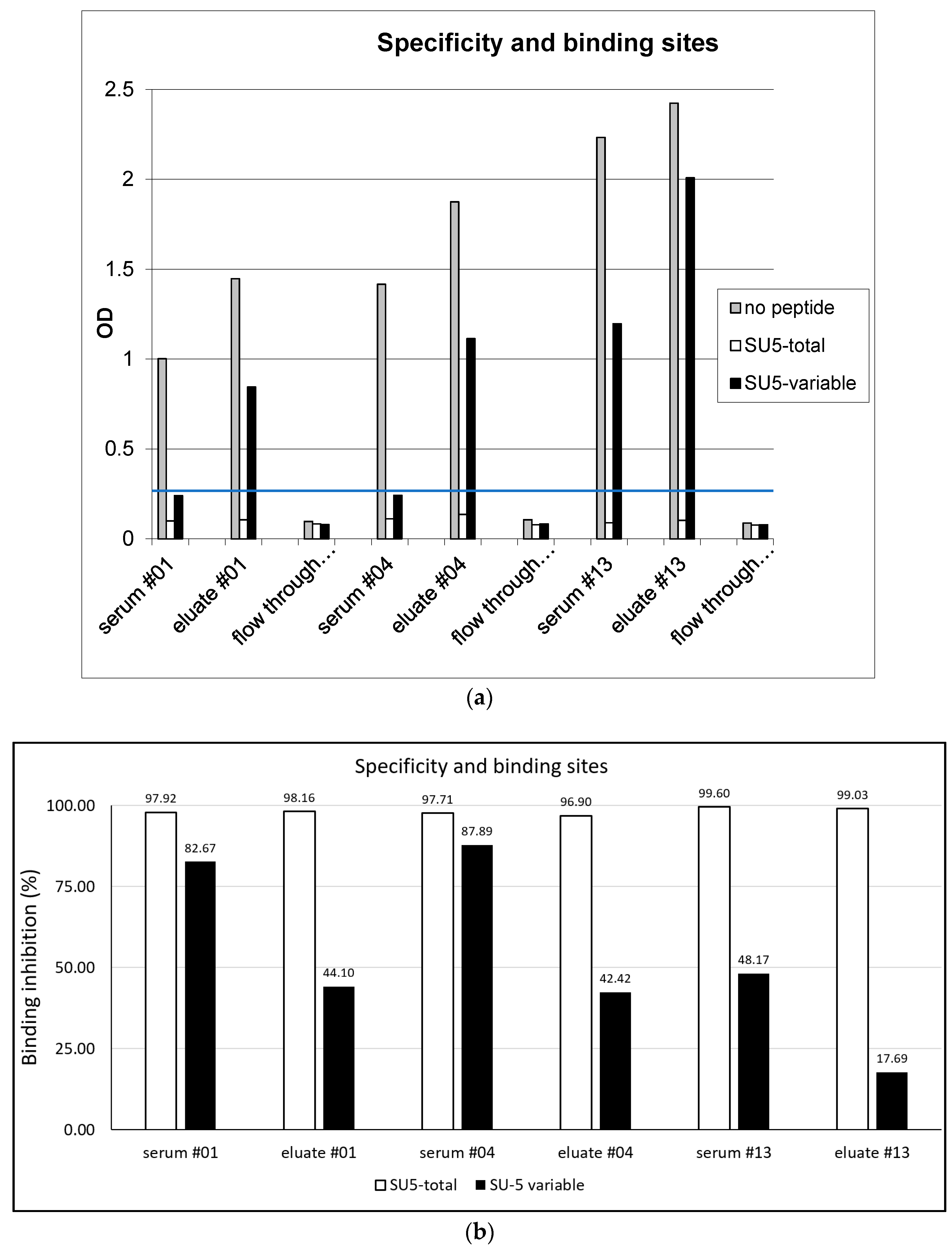
3.2. Binding Specificity of Anti-SU5 Antibody
3.3. Neutralizing Activity
3.4. Binding of Anti-SU5 Antibody to Shed Envelope Glycoproteins
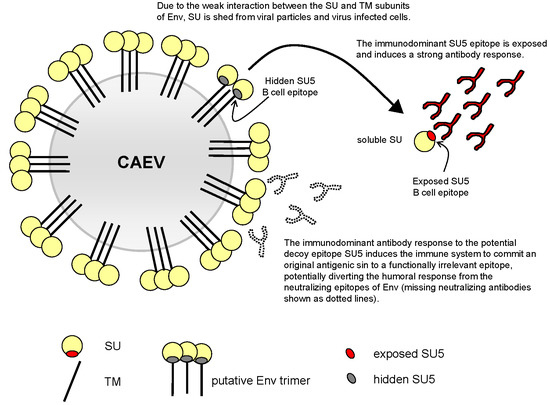
3.5. Model of SU5 Availability to B Cells
3.6. Analysis of SU-5 Sequences in a Particular Epidemiological Unit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Leroux, C.; Cruz, J.C.; Mornex, J.F. Srlvs: A genetic continuum of lentiviral species in sheep and goats with cumulative evidence of cross species transmission. Curr. HIV Res. 2010, 8, 94–100. [Google Scholar] [PubMed]
- Minardi da Cruz, J.C.; Singh, D.K.; Lamara, A.; Chebloune, Y. Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range. Viruses 2013, 5, 1867–1884. [Google Scholar] [CrossRef] [PubMed]
- Blacklaws, B.A. Small ruminant lentiviruses: Immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G.; Blatti-Cardinaux, L. Small ruminant lentivirus infections in goats. In Recent Adavances in Goat Diseases; Tempesta, M., Ed.; International Veterinary Information Service: Ithaca, NY, USA, 2016. [Google Scholar]
- Knowles, D.P.; Cheevers, W.P.; McGuire, T.C.; Stem, T.A.; Gorham, J.R. Severity of arthritis is predicted by antibody response to gp135 in chronic infection with caprine arthritis- encephalitis virus. J. Virol. 1990, 64, 2396–2398. [Google Scholar] [PubMed]
- Huso, D.L.; Narayan, O.; Hart, G.W. Sialic acid on the surface of caprine arthritis-encephalitis virus define the biological properties of the virus. J. Virol. 1988, 62, 1974–1980. [Google Scholar] [PubMed]
- Valas, S.; Benoit, C.; Baudry, G.; Perrin, G.; Mamoun, R.Z. Variability and immunogenicity of caprine arthritis-encephalitis virus surface glycoprotein. J. Virol. 2000, 74, 6178–6185. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G.; Hertig, C.; Zahno, M.L.; Vogt, H.-R.; Dufour, S.; Cordano, P.; Peterhans, E.; Cheevers, W.P.; Sonigo, P.; Pancino, G. B-cell epitopes of the envelope glycoprotein of caprine arthritis-encephalitis virus and antibody response in infected goats. J. Gen. Virol. 2000, 81, 2929–2940. [Google Scholar] [CrossRef] [PubMed]
- Mordasini, F.; Vogt, H.R.; Zahno, M.L.; Maeschli, A.; Nenci, C.; Zanoni, R.; Peterhans, E.; Bertoni, G. Analysis of the antibody response to an immunodominant epitope of the envelope glycoprotein of a lentivirus and its diagnostic potential. J. Clin. Microbiol. 2006, 44, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G.; Cardinaux, L.; Deubelbeiss, M.; Zahno, M.-L.; Vogt, H.-R. SU5 serology as a novel tool to support a challenging caprine arthritis encephalitis virus (CAEV) eradication campaign. In Lbh: 7. Leipziger Tierärztekongress; Rackwitz, R., Pees, M., Aschenbach, J.R., Gäbel, G., Eds.; University of Leipzig: Leipzig, Germany, 2014; pp. 229–232. [Google Scholar]
- Haflidadottir, B.S.; Matthiasdottir, S.; Agnarsdottir, G.; Torsteinsdottir, S.; Petursson, G.; Andresson, O.S.; Andresdottir, V. Mutational analysis of a principal neutralization domain of visna/maedi virus envelope glycoprotein. J. Gen. Virol. 2008, 89, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Andresdottir, V.; Skraban, R.; Matthiasdottir, S.; Lutley, R.; Agnarsdottir, G.; Thorsteinsdottir, H. Selection of antigenic variants in maedi-visna virus infection. J. Gen. Virol. 2002, 83, 2543–2551. [Google Scholar] [CrossRef] [PubMed]
- Pyper, J.M.; Clements, J.E.; Gonda, M.A.; Narayan, O. Sequence homology between cloned caprine arthritis encephalitis virus and visna virus, two neurotropic lentiviruses. J. Virol. 1986, 58, 665–700. [Google Scholar] [PubMed]
- Ravazzolo, A.P.; Nenci, C.; Vogt, H.-R.; Waldvogel, A.; Obexer-Ruff, G.; Peterhans, E.; Bertoni, G. Viral load, organ distribution, histopathological lesions, and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology 2006, 350, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Saltarelli, M.; Querat, G.; Konings, D.A.; Vigne, R.; Clements, J.E. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology 1990, 179, 347–364. [Google Scholar] [CrossRef]
- Bertoni, G.; Zahno, M.-L.; Zanoni, R.; Vogt, H.-R.; Peterhans, E.; Ruff, G.; Cheevers, W.P.; Sonigo, P.; Pancino, G. Antibody reactivity to the immunodominant epitopes of the caprine arthritis-encephalitis virus gp38 transmembrane protein associates with the development of arthritis. J. Virol. 1994, 68, 7139–7147. [Google Scholar] [PubMed]
- Perry, L.L.; Wilkerson, M.J.; Hullinger, G.A.; Cheevers, W.P. Depressed CD4+ T lymphocyte proliferative response and enhanced antibody response to viral antigen in chronic lentivirus- induced arthritis. J. Infect. Dis. 1995, 171, 328–334. [Google Scholar] [CrossRef] [PubMed]
- McGuire, T.C.; Norton, L.K.; O’Rourke, K.I.; Cheevers, W.P. Antigenic variation of neutralization-sensitive epitopes of caprine arthritis-encephalitis lentivirus during persistent arthritis. J. Virol. 1988, 62, 3488–3492. [Google Scholar] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Cardinaux, L.; Zahno, M.L.; Deubelbeiss, M.; Zanoni, R.; Vogt, H.R.; Bertoni, G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet. Microbiol. 2013, 162, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Skraban, R.; Matthiasdottir, S.; Torsteinsdottir, S.; Agnarsdottir, G.; Gudmundsson, B.; Georgsson, G.; Meloen, R.H.; Andresson, O.S.; Staskus, K.A.; Thormar, H.; et al. Naturally occurring mutations within 39 amino acids in the envelope glycoprotein of maedi-visna virus alter the neutralization phenotype. J. Virol. 1999, 73, 8064–8072. [Google Scholar] [PubMed]
- West, A.P., Jr.; Scharf, L.; Scheid, J.F.; Klein, F.; Bjorkman, P.J.; Nussenzweig, M.C. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 2014, 156, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Pantophlet, R.; Burton, D.R. Gp120: Target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 2006, 24, 739–769. [Google Scholar] [CrossRef] [PubMed]
- Garrity, R.R.; Rimmelzwaan, G.; Minassian, A.; Tsai, W.P.; Lin, G.; de Jong, J.J.; Goudsmit, J.; Nara, P.L. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 1997, 159, 279–289. [Google Scholar] [PubMed]
- Parren, P.W.; Burton, D.R. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 2001, 77, 195–262. [Google Scholar] [PubMed]
- Chebloune, Y.; Sheffer, D.; Karr, B.M.; Stephens, E.; Narayan, O. Restrictive type of replication of ovine/caprine lentiviruses in ovine fibroblast cell cultures. Virology 1996, 222, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Knowles, D.P.; Cheevers, W.P.; McGuire, T.C.; Brassfield, A.L.; Harwood, W.G.; Stem, T.A. Structure and genetic variability of envelope glycoproteins of two antigenic variants of caprine arthritis-encephalitis lentivirus. J. Virol. 1991, 65, 5744–5750. [Google Scholar] [PubMed]
- Hotzel, I.; Cheevers, W.P. Mutations increasing exposure of a receptor binding site epitope in the soluble and oligomeric forms of the caprine arthritis-encephalitis lentivirus envelope glycoprotein. Virology 2005, 339, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Hotzel, I.; Cheevers, W.P. Caprine arthritis-encephalitis virus envelope surface glycoprotein regions interacting with the transmembrane glycoprotein: Structural and functional parallels with human immunodeficiency virus type 1 gp120. J. Virol. 2003, 77, 11578–11587. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J.D.; Kumpula-McWhirter, N.M.; Hotzel, K.J.; Gonzalez, M.; Cheevers, W.P. Glycosylation of immunodominant linear epitopes in the carboxy-terminal region of the caprine arthritis-encephalitis virus surface envelope enhances vaccine-induced type-specific and cross-reactive neutralizing antibody responses. J. Virol. 2004, 78, 9190–9202. [Google Scholar] [CrossRef] [PubMed]
- Davenport, F.M.; Hennessy, A.V. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J. Exp. Med. 1956, 104, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Parren, P.I.; Burton, D.R.; Sattentau, Q.J. HIV-1 antibody—Debris or virion? Nat. Med. 1997, 3, 366–367. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Park, C.; Cho, S.H.; Chung, J. The level of decoy epitope in PCV2 vaccine affects the neutralizing activity of sera in the immunized animals. Biochem. Biophys. Res. Commun. 2018, 496, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, X.; Liu, J.; Diao, W.; Zhang, L.; Xiao, Y.; Wei, H.; Yu, Y.; Yu, Y.; Wang, L. Replacing the decoy epitope of PCV2b capsid protein with a protective epitope enhances efficacy of PCV2b vaccine. Vaccine 2016, 34, 6358–6366. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.E.; Huso, D.; Hart, G.; Narayan, O. Modulation of lentivirus replication by antibodies. Non-neutralizing antibodies to caprine arthritis-encephalitis virus enhance early stages of infection in macrophages, but do not cause increased production of virions. J. Gen. Virol. 1989, 70, 2221–2226. [Google Scholar] [CrossRef] [PubMed]
- Hachimura, S.; Enomoto, A.; Kaminogawa, S. Relative positioning of the T cell and B cell determinants on an immunogenic peptide: Its influence on antibody response. Biochem. Biophys. Res. Commun. 1990, 169, 803–808. [Google Scholar] [CrossRef]
- Nenci, C.; Zahno, M.L.; Vogt, H.-R.; Obexer-Ruff, G.; Doherr, M.G.; Zanoni, R.; Peterhans, E.; Bertoni, G. Vaccination with a T-cell-priming Gag peptide of caprine arthritis encephalitis virus enhances virus replication transiently in vivo. J. Gen. Virol. 2007, 88, 1589–1593. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahno, M.-L.; Bertoni, G. An Immunodominant Region of the Envelope Glycoprotein of Small Ruminant Lentiviruses May Function as Decoy Antigen. Viruses 2018, 10, 231. https://doi.org/10.3390/v10050231
Zahno M-L, Bertoni G. An Immunodominant Region of the Envelope Glycoprotein of Small Ruminant Lentiviruses May Function as Decoy Antigen. Viruses. 2018; 10(5):231. https://doi.org/10.3390/v10050231
Chicago/Turabian StyleZahno, Marie-Luise, and Giuseppe Bertoni. 2018. "An Immunodominant Region of the Envelope Glycoprotein of Small Ruminant Lentiviruses May Function as Decoy Antigen" Viruses 10, no. 5: 231. https://doi.org/10.3390/v10050231