Glycosylation of Hemagglutinin and Neuraminidase of Influenza A Virus as Signature for Ecological Spillover and Adaptation among Influenza Reservoirs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Prediction of Potential N-Glycosylation Sites in HA and NA of Influenza A H1N1 Viruses
2.2. Phylogenetic Tree
2.3. Homology Modeling, in Silico Protein Glycosylation, and Visualization
2.4. Structural Analysis
3. Results
3.1. Dynamics of N-Linked Glycosylations in the HA and NA of Influenza H1N1 Viruses
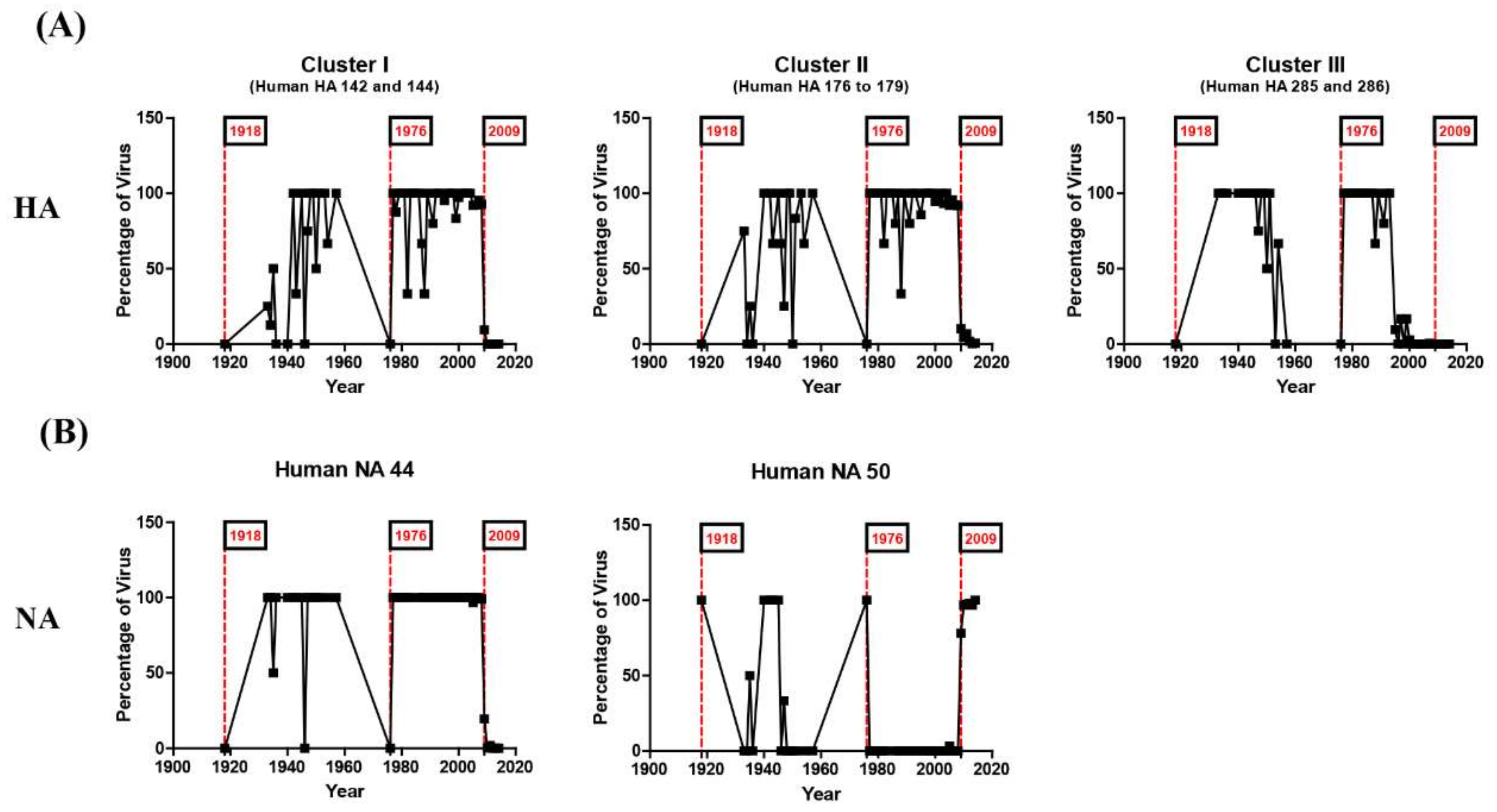
3.2. N-Linked Glycosylations as a Predictive Signature of Influenza Pandemics
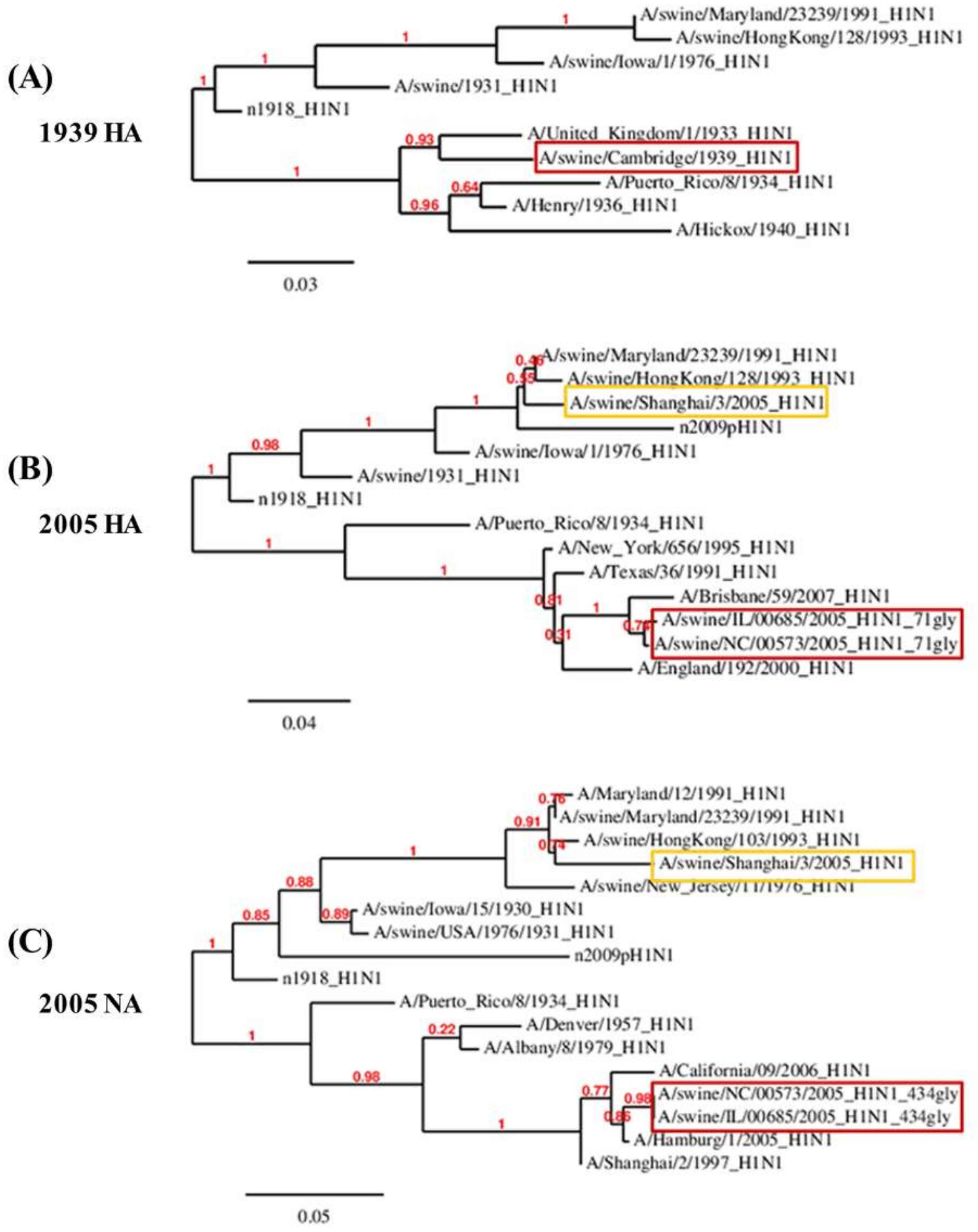
3.3. Reverse Zoonosis of Influenza Viruses Is Traceable by N-Linked Glycosylation
3.4. N-Linked Glycosylation in Human Influenza H1N1 Viruses Emerged after the 2009 Pandemic
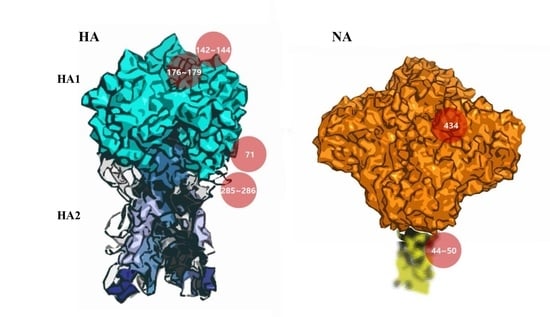
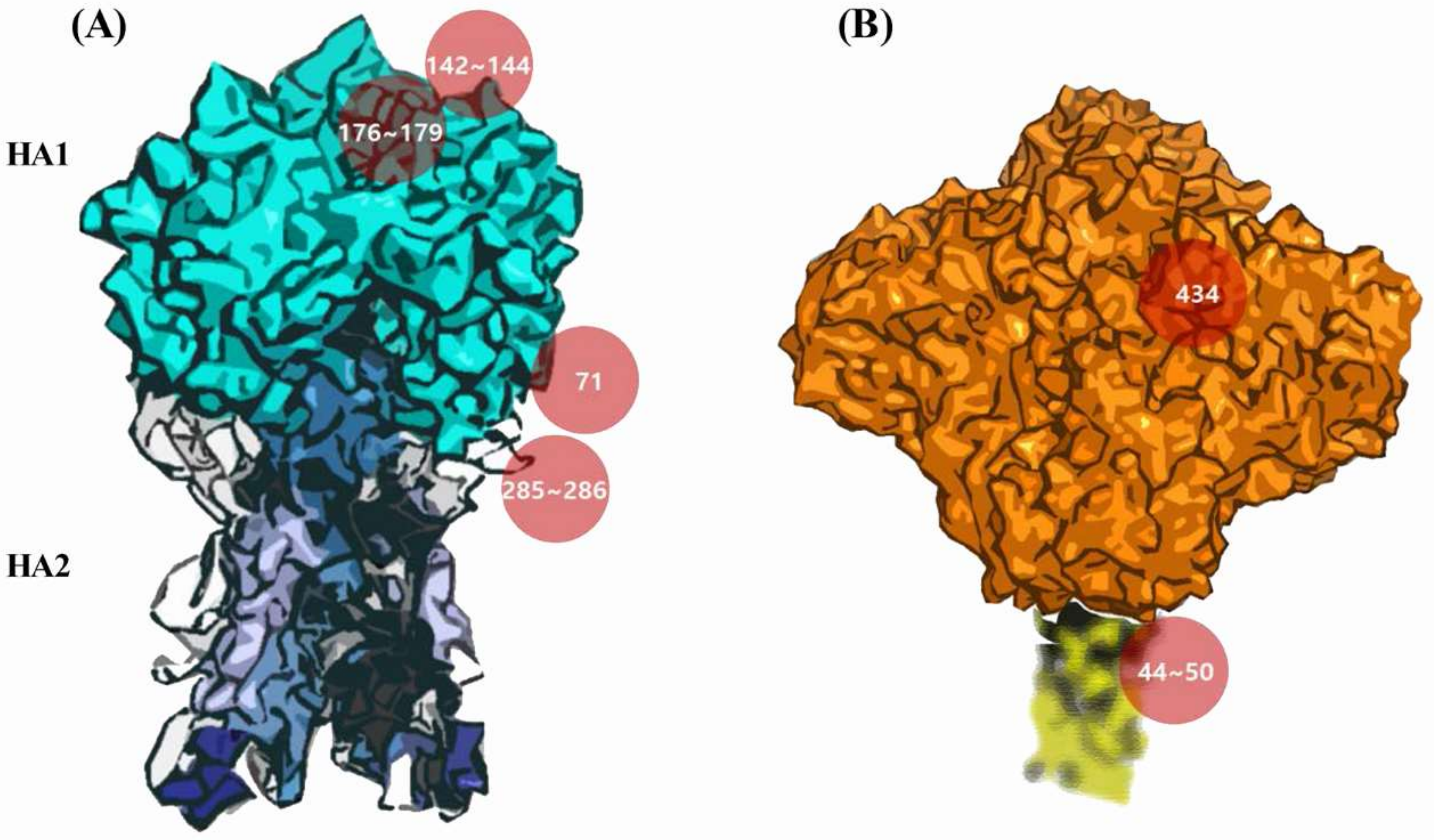
3.5. Potential Antigenic Shielding by N-Linked Glycosylation
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Glezen, W.P. Clinical practice. Prevention and treatment of seasonal influenza. N. Engl. J. Med. 2008, 359, 2579–2585. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.W. A history of influenza. J. Appl. Microbiol. 2001, 91, 572–579. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014, 12, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.C.; Garten, W.; Klenk, H.D. Role of conserved glycosylation sites in maturation and transport of influenza a virus hemagglutinin. J. Virol. 1993, 67, 3048–3060. [Google Scholar] [PubMed]
- Gallagher, P.J.; Henneberry, J.M.; Sambrook, J.F.; Gething, M.J. Glycosylation requirements for intracellular transport and function of the hemagglutinin of influenza virus. J. Virol. 1992, 66, 7136–7145. [Google Scholar] [PubMed]
- Klenk, H.D.; Wagner, R.; Heuer, D.; Wolff, T. Importance of hemagglutinin glycosylation for the biological functions of influenza virus. Virus Res. 2002, 82, 73–75. [Google Scholar] [CrossRef]
- Job, E.R.; Deng, Y.M.; Barfod, K.K.; Tate, M.D.; Caldwell, N.; Reddiex, S.; Maurer-Stroh, S.; Brooks, A.G.; Reading, P.C. Addition of glycosylation to influenza a virus hemagglutinin modulates antibody-mediated recognition of H1N1 2009 pandemic viruses. J. Immunol. 2013, 190, 2169–2177. [Google Scholar] [CrossRef] [PubMed]
- Wanzeck, K.; Boyd, K.L.; McCullers, J.A. Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am. J. Respir. Crit. Care Med. 2011, 183, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Suzuki, Y. Evidence for N-glycan shielding of antigenic sites during evolution of human influenza a virus hemagglutinin. J. Virol. 2012, 86, 3446–3451. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Job, E.R.; Deng, Y.M.; Gunalan, V.; Maurer-Stroh, S.; Reading, P.C. Playing hide and seek: How glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014, 6, 1294–1316. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.J.; Boyington, J.C.; Dai, K.; Houser, K.V.; Pearce, M.B.; Kong, W.P.; Yang, Z.Y.; Tumpey, T.M.; Nabel, G.J. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010, 2, 24ra21. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Brooks, A.G.; Reading, P.C. Specific sites of N-linked glycosylation on the hemagglutinin of H1N1 subtype influenza a virus determine sensitivity to inhibitors of the innate immune system and virulence in mice. J. Immunol. 2011, 187, 1884–1894. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, Q.; Zhao, F.; Chen, W.; Li, Z. Prediction of biological functions on glycosylation site migrations in human influenza H1N1 viruses. PLoS ONE 2012, 7, e32119. [Google Scholar] [CrossRef] [PubMed]
- Sencer, D.J.; Millar, J.D. Reflections on the 1976 swine flu vaccination program. Emerg. Infect. Dis. 2006, 12, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Sencer, D.J. Perspective: Swine-origin influenza: 1976 and 2009. Clin. Infect. Dis. 2011, 52 (Suppl. S1), S4–S7. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Cauchemez, S.; Hayden, F.G. “Prepandemic” immunization for novel influenza viruses, “swine flu” vaccine, guillain-barre syndrome, and the detection of rare severe adverse events. J. Infect. Dis. 2009, 200, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, Q.; Zhao, F.; Chen, W.; Li, Z. Glycosylation site alteration in the evolution of influenza a (H1N1) viruses. PLoS ONE 2011, 6, e22844. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Chen, J.R.; Tseng, Y.C.; Hsu, C.H.; Hung, Y.F.; Chen, S.W.; Chen, C.M.; Khoo, K.H.; Cheng, T.J.; Cheng, Y.S.; et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. USA 2009, 106, 18137–18142. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.B.; Husain, M. Reemergence of swine flu H1N1 in india: First outbreak of 2015. Austin J. Infect. Dis. 2015, 2, 1015. [Google Scholar]
- Tharakaraman, K.; Sasisekharan, R. Influenza surveillance: 2014–2015 H1N1 “swine”-derived influenza viruses from India. Cell Host Microbe 2015, 17, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 310–322. [Google Scholar] [CrossRef]
- Joshi, H.J.; Gupta, R. Eukaryotic glycosylation: Online methods for site prediction on protein sequences. Methods Mol. Biol. 2015, 1273, 127–137. [Google Scholar] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Yang, S.; Guo, Y.; Yang, L.; Bai, T.; Yu, Z.; Li, X.; Li, M.; Guo, J.; Wang, D.; et al. Imported pigs may have introduced the first classical swine influenza viruses into mainland China. Infect. Genet. Evol. 2013, 17, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with swiss-model and swiss-pdbviewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. S1), S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. Swiss-model: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Ekiert, D.C.; Krause, J.C.; Hai, R.; Crowe, J.E., Jr.; Wilson, I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010, 328, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Stray, S.J.; Pittman, L.B. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol. J. 2012, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Colman, P.M.; Varghese, J.N.; Laver, W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983, 303, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Shrake, A.; Rupley, J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973, 79, 351–371. [Google Scholar] [CrossRef]
- Jung, J.S.; Lee, B.K. Protein structure alignment using environmental profiles. Protein Eng. 2000, 13, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.H.; Taubenberger, J.K.; Fanning, T.G. Evidence of an absence: The genetic origins of the 1918 pandemic influenza virus. Nat. Rev. Microbiol. 2004, 2, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza a epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Zhong, G.; Russell, C.A.; Nakajima, N.; Hatta, M.; Hanson, A.; McBride, R.; Burke, D.F.; Takahashi, K.; Fukuyama, S.; et al. Circulating avian influenza viruses closely related to the 1918 virus have pandemic potential. Cell Host Microbe 2014, 15, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Neumeier, E.; Meier-Ewert, H. Nucleotide sequence analysis of the HA1 coding portion of the haemagglutinin gene of swine H1N1 influenza viruses. Virus Res. 1992, 23, 107–117. [Google Scholar] [CrossRef]
- Medina, R.A.; Stertz, S.; Manicassamy, B.; Zimmermann, P.; Sun, X.; Albrecht, R.A.; Uusi-Kerttula, H.; Zagordi, O.; Belshe, R.B.; Frey, S.E.; et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013, 5, 187ra170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovida, F.; Piralla, A.; Marzani, F.C.; Moreno, A.; Campanini, G.; Mojoli, F.; Pozzi, M.; Girello, A.; Chiapponi, C.; Vezzoli, F.; et al. Swine influenza a (H1N1) virus (siv) infection requiring extracorporeal life support in an immunocompetent adult patient with indirect exposure to pigs, Italy, October 2016. Euro Surveill. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Gamblin, S.J.; Haire, L.F.; Russell, R.J.; Stevens, D.J.; Xiao, B.; Ha, Y.; Vasisht, N.; Steinhauer, D.A.; Daniels, R.S.; Elliot, A.; et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 2004, 303, 1838–1842. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; McBride, R.; Nycholat, C.M.; Paulson, J.C.; Wilson, I.A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 2012, 86, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Tsibane, T.; Krammer, F.; Hai, R.; Rahmat, S.; Basler, C.F.; Palese, P. 1976 and 2009 H1N1 influenza virus vaccines boost anti-hemagglutinin stalk antibodies in humans. J. Infect. Dis. 2013, 207, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Schmeisser, F.; Friedman, R.; Besho, J.; Lugovtsev, V.; Soto, J.; Wang, W.; Weiss, C.; Williams, O.; Xie, H.; Ye, Z.; et al. Neutralizing and protective epitopes of the 2009 pandemic influenza H1N1 hemagglutinin. Influenza Other Respir. Viruses 2013, 7, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Das, S.R.; Hensley, S.E.; David, A.; Schmidt, L.; Gibbs, J.S.; Puigbo, P.; Ince, W.L.; Bennink, J.R.; Yewdell, J.W. Fitness costs limit influenza a virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. USA 2011, 108, E1417–E1422. [Google Scholar] [CrossRef] [PubMed]
- Kosik, I.; Ince, W.L.; Gentles, L.E.; Oler, A.J.; Kosikova, M.; Angel, M.; Magadan, J.G.; Xie, H.; Brooke, C.B.; Yewdell, J.W. Influenza a virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018, 14, e1006796. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Vincent, A.L. Reverse zoonosis of influenza to swine: New perspectives on the human-animal interface. Trends Microbiol. 2015, 23, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Messenger, A.M.; Barnes, A.N.; Gray, G.C. Reverse zoonotic disease transmission (zooanthroponosis): A systematic review of seldom-documented human biological threats to animals. PLoS ONE 2014, 9, e89055. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.; Markowska-Daniel, I. Phylogenetic evolution of swine-origin human influenza virus: A pandemic H1N1 2009. Pol. J. Vet. Sci. 2010, 13, 491–500. [Google Scholar] [PubMed]
- Newman, A.P.; Reisdorf, E.; Beinemann, J.; Uyeki, T.M.; Balish, A.; Shu, B.; Lindstrom, S.; Achenbach, J.; Smith, C.; Davis, J.P. Human case of swine influenza a (H1N1) triple reassortant virus infection, wisconsin. Emerg. Infect. Dis. 2008, 14, 1470–1472. [Google Scholar] [CrossRef] [PubMed]
- Russell, K.; Blanton, L.; Kniss, K.; Mustaquim, D.; Smith, S.; Cohen, J.; Garg, S.; Flannery, B.; Fry, A.M.; Grohskopf, L.A.; et al. Update: Influenza activity--united states, October 4, 2015–February 6, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Gaymard, A.; Le Briand, N.; Frobert, E.; Lina, B.; Escuret, V. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin. Microbiol. Infect. 2016, 22, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Evans, J.G.; Stambas, J. Influenza reverse genetics: Dissecting immunity and pathogenesis. Expert Rev. Mol. Med. 2014, 16, e2. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Ozawa, M.; Kawaoka, Y. Reverse genetics of influenza viruses. Methods Mol. Biol. 2012, 865, 193–206. [Google Scholar] [PubMed]


| (A) HA Glycosylation | |||||||
|---|---|---|---|---|---|---|---|
| +142 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| Sa | 141 | P | 91 | 78 | 58 | 47 | |
| Sa | 142 | N | 73 | 33 | 69 | 17 | |
| +177 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| Sa | 177 | K | 67 | 16 | 87 | 43 | |
| Sb | 201 | T | 45 | 39 | 83 | 69 | |
| Sb | 202 | S | 68 | 3 | 82 | 33 | |
| Sb | 206 | Q | 54 | 25 | 83 | 59 | |
| Sb | 212 | A | 58 | 0 | 83 | 24 | |
| +286 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| H1C | 286 | D | 76 | 32 | 85 | 47 | |
| +71 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| - | - | - | - | - | - | - | |
| +212 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant Pol (%) |
| Sb | 202 | S | 68 | 22 | 82 | 36 | |
| Sb | 206 | Q | 54 | 37 | 83 | 71 | |
| Sb | 211 | N | 38 | 1 | 70 | 33 | |
| Sb | 212 | A | 58 | 8 | 83 | 36 | |
| (B) NA Glycosylation | |||||||
| +434 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| AS5 | 429 | G | 0 | 0 | 43 | 56 | |
| +341 | Antigenic Site | Position | WT AA | WT acc (%) | Mutant acc (%) | WT pol (%) | Mutant pol (%) |
| AS5 | 430 | T | 37 | 25 | 73 | 66 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, P.; Jang, Y.H.; Kwon, S.B.; Lee, C.M.; Han, G.; Seong, B.L. Glycosylation of Hemagglutinin and Neuraminidase of Influenza A Virus as Signature for Ecological Spillover and Adaptation among Influenza Reservoirs. Viruses 2018, 10, 183. https://doi.org/10.3390/v10040183
Kim P, Jang YH, Kwon SB, Lee CM, Han G, Seong BL. Glycosylation of Hemagglutinin and Neuraminidase of Influenza A Virus as Signature for Ecological Spillover and Adaptation among Influenza Reservoirs. Viruses. 2018; 10(4):183. https://doi.org/10.3390/v10040183
Chicago/Turabian StyleKim, Paul, Yo Han Jang, Soon Bin Kwon, Chung Min Lee, Gyoonhee Han, and Baik Lin Seong. 2018. "Glycosylation of Hemagglutinin and Neuraminidase of Influenza A Virus as Signature for Ecological Spillover and Adaptation among Influenza Reservoirs" Viruses 10, no. 4: 183. https://doi.org/10.3390/v10040183