The French Armed Forces Virology Unit: A Chronological Record of Ongoing Research on Orthopoxvirus
Abstract
:1. Introduction
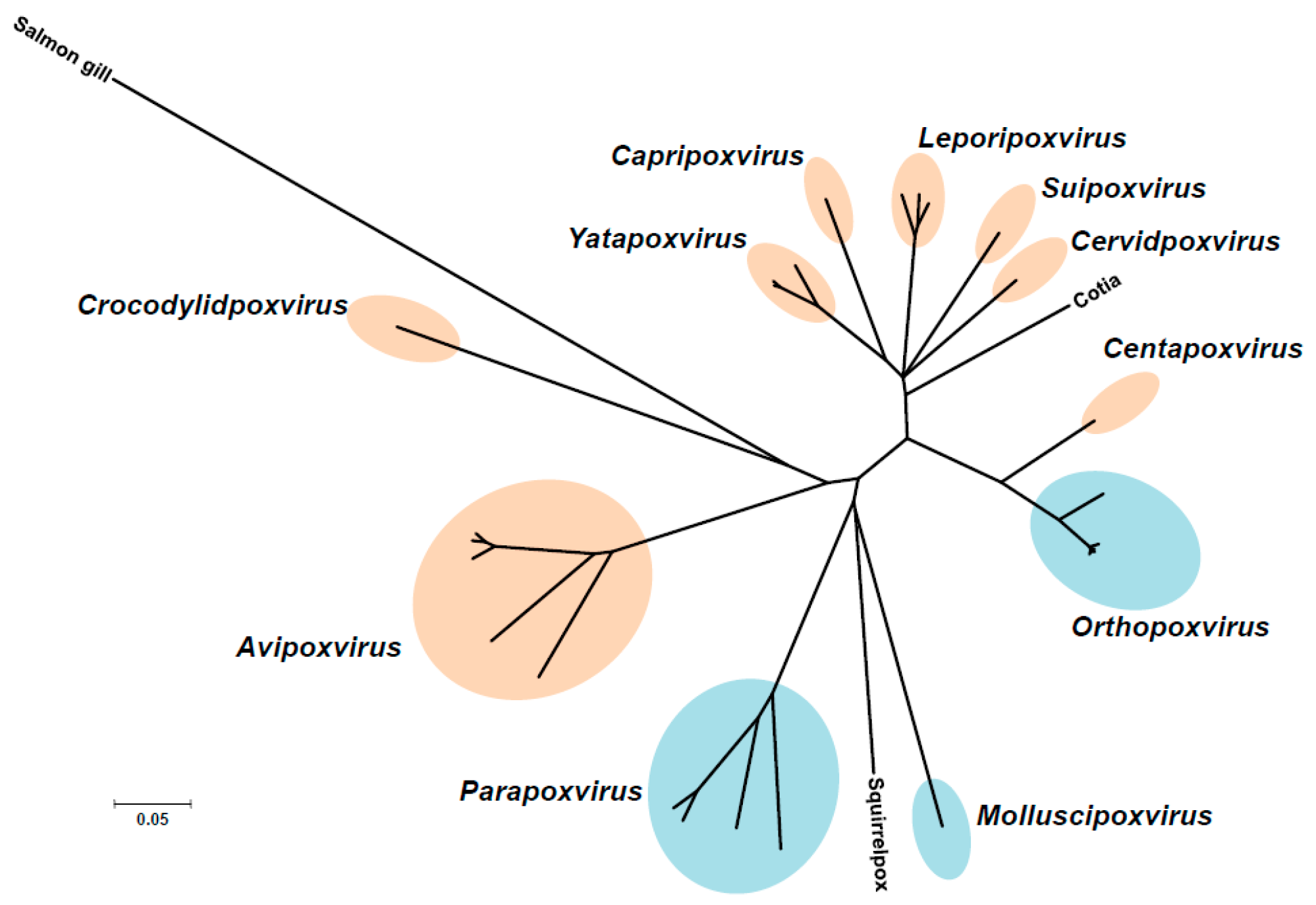
2. The Poxviruses
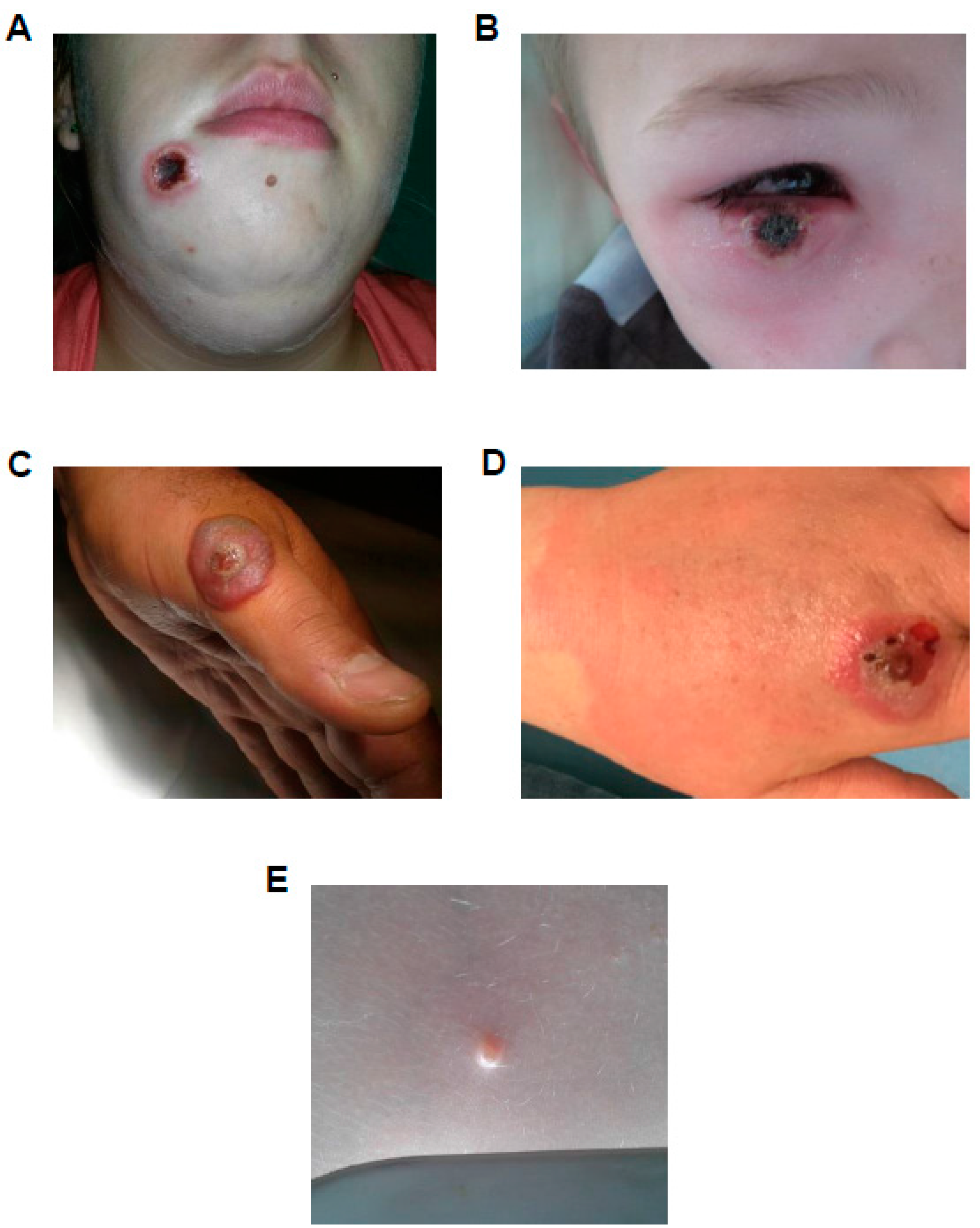
3. French National Reference Center-Expert Laboratory for Orthopoxvirus
4. Smallpox Vaccine, Past and Present
5. Antivirals
6. Conclusions
Conflicts of Interest
References
- World Health Assembly. Global Eradication of Smallpox; World Health Assembly: Geneva, Switzerland, 1980. [Google Scholar]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The detection of monkeypox in humans in the western hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.; Hughes, C.M.; Kabamba, J.; Malekani, J.; et al. Extended human-to-human transmission during a monkeypox outbreak in the democratic republic of the Congo. Emerg. Infect. Dis. 2016, 22, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Abrahao, J.S.; Campos, R.K.; Trindade Gde, S.; Guimaraes da Fonseca, F.; Ferreira, P.C.; Kroon, E.G. Outbreak of severe zoonotic vaccinia virus infection, southeastern Brazil. Emerg. Infect. Dis. 2015, 21, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Scaramozzino, N.; Ferrier-Rembert, A.; Favier, A.L.; Rothlisberger, C.; Richard, S.; Crance, J.M.; Meyer, H.; Garin, D. Real-time PCR to identify variola virus or other human pathogenic orthopox viruses. Clin. Chem. 2007, 53, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.L.; Peng, C.; McFadden, G.; Rothenburg, S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014, 21, 15–40. [Google Scholar] [CrossRef] [PubMed]
- Bessaud, M.; Ferraris, O.; Ferrier-Rembert, A.; Jarjaval, F.; Favier, A.; Iseni, F.; Peyrefitte, C. Les parapoxvirus, des agents zoonotiques encore mal connus. Virologie 2016, 20, 218–230. [Google Scholar]
- Nishiura, H.; Schwehm, M.; Eichner, M. Still protected against smallpox? Estimation of the duration of vaccine-induced immunity against smallpox. Epidemiology 2006, 17, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Demkowicz, W.E., Jr.; Littaua, R.A.; Wang, J.; Ennis, F.A. Human cytotoxic T-cell memory: Long-lived responses to vaccinia virus. J. Virol. 1996, 70, 2627–2631. [Google Scholar] [PubMed]
- Beeching, N.J.; Dance, D.A.; Miller, A.R.; Spencer, R.C. Biological warfare and bioterrorism. BMJ 2002, 324, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Huhn, G.D.; Chase, R.A.; Dworkin, M.S. Monkeypox in the western hemisphere. N. Engl. J. Med. 2004, 350, 1790–1791. [Google Scholar] [PubMed]
- Brochard, J.; Guimard, T.; de Bataille, S.; Poiraud, C.; Ferraris, O. An atypical necrotic wound. Med. Mal. Infect. 2015, 45, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Ducournau, C.; Ferrier-Rembert, A.; Ferraris, O.; Joffre, A.; Favier, A.L.; Flusin, O.; van Cauteren, D.; Kecir, K.; Auburtin, B.; Vedy, S.; et al. Concomitant human infections with 2 cowpox virus strains in related cases, france, 2011. Emerg. Infect. Dis. 2013, 19, 1996–1999. [Google Scholar] [CrossRef] [PubMed]
- Duraffour, S.; Ferraris, O.; Peyrefitte, C. Cowpox et monkeypox. EMC Mal. Infect. 2016, 8-050-M-10. [Google Scholar] [CrossRef]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Buttner, M.; et al. Cowpox virus transmission from pet rats to humans, germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Ninove, L.; Domart, Y.; Vervel, C.; Voinot, C.; Salez, N.; Raoult, D.; Meyer, H.; Capek, I.; Zandotti, C.; Charrel, R.N. Cowpox virus transmission from pet rats to humans, france. Emerg. Infect. Dis. 2009, 15, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Hanson, D.; Diven, D.G. Molluscum contagiosum. Dermatol. Online J. 2003, 9, 2. [Google Scholar] [PubMed]
- Garin, D.; Crance, J.M.; Fuchs, F.; Autran, B.; Drillien, R. Update on smallpox vaccines. Med. Mal. Infect. 2004, 34, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.M.; Ruben, F.L.; Neff, J.M.; Millar, J.D. Complications of smallpox vaccination, 1968: Results of ten statewide surveys. J. Infect. Dis. 1970, 122, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.M.; Ruben, F.L.; Neff, J.M.; Millar, J.D. Complications of smallpox vaccination, 1968. N. Engl. J. Med. 1969, 281, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Contraindications to Vaccination. Available online: https://www.Cdc.Gov/smallpox/clinicians/vaccination-contraindications1.Html (5 December 2016).
- Crance, J.M.L.D.; Scaramozzino, N.; Ferrier-Rembert, A.; Agay, D.; Garin, D. Variole: La menace fantôme. Méd. Armées 2002, 30, 495–502. [Google Scholar]
- Bricaire, F.; Combadiere, B.; Rouleau, E.; Bossi, P.; Garin, D.; Lebrun-Vignes, B.; Autran, B. Evaluation of residual immune response against human pox virus before and after revaccination in healthy volunteers. Bull. Acad. Natl. Med. 2006, 190, 1035–1046. [Google Scholar] [PubMed]
- Appel D’Offre Organisation JNI 2021–2024. Available online: http://www.Infectiologie.Com/userfiles/file/medias/_documents/bt/vaccin%20variole.Pdf (accessed on 12–14 March 2003).
- Ferrier-Rembert, A.; Drillien, R.; Meignier, B.; Garin, D.; Crance, J.M. Safety, immunogenicity and protective efficacy in mice of a new cell-cultured lister smallpox vaccine candidate. Vaccine 2007, 25, 8290–8297. [Google Scholar] [CrossRef] [PubMed]
- Ferrier-Rembert, A.; Drillien, R.; Tournier, J.N.; Garin, D.; Crance, J.M. Intranasal cowpox virus infection of the mouse as a model for preclinical evaluation of smallpox vaccines. Vaccine 2007, 25, 4809–4817. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, J.; Perkus, M.E.; Taylor, J.; Norton, E.K.; Audonnet, J.C.; Cox, W.I.; Davis, S.W.; van der Hoeven, J.; Meignier, B.; Riviere, M.; et al. Nyvac: A highly attenuated strain of vaccinia virus. Virology 1992, 188, 217–232. [Google Scholar] [CrossRef]
- Ferrier-Rembert, A.; Drillien, R.; Tournier, J.N.; Garin, D.; Crance, J.M. Short- and long-term immunogenicity and protection induced by non-replicating smallpox vaccine candidates in mice and comparison with the traditional 1st generation vaccine. Vaccine 2008, 26, 1794–1804. [Google Scholar] [CrossRef] [PubMed]
- Garcel, A.; Crance, J.M.; Drillien, R.; Garin, D.; Favier, A.L. Genomic sequence of a clonal isolate of the vaccinia virus lister strain employed for smallpox vaccination in france and its comparison to other orthopoxviruses. J. Gen. Virol. 2007, 88, 1906–1916. [Google Scholar] [CrossRef] [PubMed]
- Dimier, J.; Ferrier-Rembert, A.; Pradeau-Aubreton, K.; Hebben, M.; Spehner, D.; Favier, A.L.; Gratier, D.; Garin, D.; Crance, J.M.; Drillien, R. Deletion of major nonessential genomic regions in the vaccinia virus lister strain enhances attenuation without altering vaccine efficacy in mice. J. Virol. 2011, 85, 5016–5026. [Google Scholar] [CrossRef] [PubMed]
- Antoine, G.; Scheiflinger, F.; Dorner, F.; Falkner, F.G. The complete genomic sequence of the modified vaccinia ankara strain: Comparison with other orthopoxviruses. Virology 1998, 244, 365–396. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Sutter, G.; Mayr, A. Mapping of deletions in the genome of the highly attenuated vaccinia virus mva and their influence on virulence. J. Gen. Virol. 1991, 72 Pt 5, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Holy, A.; Rosenberg, I.; Sakuma, T.; Balzarini, J.; Maudgal, P.C. A novel selective broad-spectrum anti-DNA virus agent. Nature 1986, 323, 464–467. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Clinical potential of the acyclic nucleoside phosphonates cidofovir, adefovir, and tenofovir in treatment of DNA virus and retrovirus infections. Clin. Microbiol. Rev. 2003, 16, 569–596. [Google Scholar] [CrossRef] [PubMed]
- Magee, W.C.; Hostetler, K.Y.; Evans, D.H. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 2005, 49, 3153–3162. [Google Scholar] [CrossRef] [PubMed]
- Prevention and Treatment. Available online: https://www.Cdc.Gov/smallpox/prevention-treatment/index.Html (accessed on 7 June 2016).
- Kern, E.R.; Hartline, C.; Harden, E.; Keith, K.; Rodriguez, N.; Beadle, J.R.; Hostetler, K.Y. Enhanced inhibition of orthopoxvirus replication in vitro by alkoxyalkyl esters of cidofovir and cyclic cidofovir. Antimicrob. Agents Chemother. 2002, 46, 991–995. [Google Scholar] [CrossRef] [PubMed]
- Olson, V.A.; Smith, S.K.; Foster, S.; Li, Y.; Lanier, E.R.; Gates, I.; Trost, L.C.; Damon, I.K. In vitro efficacy of brincidofovir against variola virus. Antimicrob. Agents Chemother. 2014, 58, 5570–5571. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.; Touchette, E.; Oberle, C.; Almond, M.; Robertson, A.; Trost, L.C.; Lampert, B.; Painter, G.; Buller, R.M. Efficacy of therapeutic intervention with an oral ether-lipid analogue of cidofovir (CMX001) in a lethal mousepox model. Antivir. Res. 2008, 77, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Trost, L.C.; Rose, M.L.; Khouri, J.; Keilholz, L.; Long, J.; Godin, S.J.; Foster, S.A. The efficacy and pharmacokinetics of brincidofovir for the treatment of lethal rabbitpox virus infection: A model of smallpox disease. Antivir. Res. 2015, 117, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Ciesla, S.L.; Trahan, J.; Wan, W.B.; Beadle, J.R.; Aldern, K.A.; Painter, G.R.; Hostetler, K.Y. Esterification of cidofovir with alkoxyalkanols increases oral bioavailability and diminishes drug accumulation in kidney. Antivir. Res. 2003, 59, 163–171. [Google Scholar] [CrossRef]
- Marty, F.M.; Winston, D.J.; Rowley, S.D.; Vance, E.; Papanicolaou, G.A.; Mullane, K.M.; Brundage, T.M.; Robertson, A.T.; Godkin, S.; Mommeja-Marin, H.; et al. CMX001 to prevent cytomegalovirus disease in hematopoietic-cell transplantation. N. Engl. J. Med. 2013, 369, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Chittick, G.; Morrison, M.; Brundage, T.; Nichols, W.G. Short-term clinical safety profile of brincidofovir: A favorable benefit-risk proposition in the treatment of smallpox. Antivir. Res. 2017, 143, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Pevear, D.C.; Davies, M.H.; Collett, M.S.; Bailey, T.; Rippen, S.; Barone, L.; Burns, C.; Rhodes, G.; Tohan, S.; et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J. Virol. 2005, 79, 13139–13149. [Google Scholar] [CrossRef] [PubMed]
- Duraffour, S.; Snoeck, R.; de Vos, R.; van Den Oord, J.J.; Crance, J.M.; Garin, D.; Hruby, D.E.; Jordan, R.; de Clercq, E.; Andrei, G. Activity of the anti-orthopoxvirus compound ST-246 against vaccinia, cowpox and camelpox viruses in cell monolayers and organotypic raft cultures. Antivir. Ther. 2007, 12, 1205–1216. [Google Scholar] [PubMed]
- Smith, S.K.; Olson, V.A.; Karem, K.L.; Jordan, R.; Hruby, D.E.; Damon, I.K. In vitro efficacy of ST246 against smallpox and monkeypox. Antimicrob. Agents Chemother. 2009, 53, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Quenelle, D.C.; Buller, R.M.; Parker, S.; Keith, K.A.; Hruby, D.E.; Jordan, R.; Kern, E.R. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob. Agents Chemother. 2007, 51, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Huggins, J.; Goff, A.; Hensley, L.; Mucker, E.; Shamblin, J.; Wlazlowski, C.; Johnson, W.; Chapman, J.; Larsen, T.; Twenhafel, N.; et al. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob. Agents Chemother. 2009, 53, 2620–2625. [Google Scholar] [CrossRef] [PubMed]
- Jordan, R.; Tien, D.; Bolken, T.C.; Jones, K.F.; Tyavanagimatt, S.R.; Strasser, J.; Frimm, A.; Corrado, M.L.; Strome, P.G.; Hruby, D.E. Single-dose safety and pharmacokinetics of ST-246, a novel orthopoxvirus egress inhibitor. Antimicrob. Agents Chemother. 2008, 52, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Chinsangaram, J.; Honeychurch, K.M.; Tyavanagimatt, S.R.; Leeds, J.M.; Bolken, T.C.; Jones, K.F.; Jordan, R.; Marbury, T.; Ruckle, J.; Mee-Lee, D.; et al. Safety and pharmacokinetics of the anti-orthopoxvirus compound ST-246 following a single daily oral dose for 14 days in human volunteers. Antimicrob. Agents Chemother. 2012, 56, 4900–4905. [Google Scholar] [CrossRef] [PubMed]
- Vora, S.; Damon, I.; Fulginiti, V.; Weber, S.G.; Kahana, M.; Stein, S.L.; Gerber, S.I.; Garcia-Houchins, S.; Lederman, E.; Hruby, D.; et al. Severe eczema vaccinatum in a household contact of a smallpox vaccinee. Clin. Infect. Dis. 2008, 46, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Jordan, R.; Leeds, J.M.; Tyavanagimatt, S.; Hruby, D.E. Development of ST-246(r) for treatment of poxvirus infections. Viruses 2010, 2, 2409–2435. [Google Scholar] [CrossRef] [PubMed]
- Challberg, M.D.; Englund, P.T. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J. Biol. Chem. 1979, 254, 7812–7819. [Google Scholar] [PubMed]
- Czarnecki, M.W.; Traktman, P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res. 2017, 234, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Upton, C.; Stuart, D.T.; McFadden, G. Identification of a poxvirus gene encoding a uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA 1993, 90, 4518–4522. [Google Scholar] [CrossRef] [PubMed]
- Stuart, D.T.; Upton, C.; Higman, M.A.; Niles, E.G.; McFadden, G. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J. Virol. 1993, 67, 2503–2512. [Google Scholar] [PubMed]
- Stanitsa, E.S.; Arps, L.; Traktman, P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 2006, 281, 3439–3451. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Klemperer, N.; Ghosh, R.; Traktman, P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 1995, 69, 5353–5361. [Google Scholar] [PubMed]
- Hutin, S.; Ling, W.L.; Round, A.; Effantin, G.; Reich, S.; Iseni, F.; Tarbouriech, N.; Schoehn, G.; Burmeister, W.P. Domain organization of vaccinia virus helicase-primase D5. J. Virol. 2016, 90, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.A.; Arps, L.; Traktman, P. Biochemical and genetic analysis of the vaccinia virus D5 protein: Multimerization-dependent atpase activity is required to support viral DNA replication. J. Virol. 2007, 81, 844–859. [Google Scholar] [CrossRef] [PubMed]
- De Silva, F.S.; Lewis, W.; Berglund, P.; Koonin, E.V.; Moss, B. Poxvirus DNA primase. Proc. Natl. Acad. Sci. USA 2007, 104, 18724–18729. [Google Scholar] [CrossRef] [PubMed]
- Sele, C.; Gabel, F.; Gutsche, I.; Ivanov, I.; Burmeister, W.P.; Iseni, F.; Tarbouriech, N. Low-resolution structure of vaccinia virus DNA replication machinery. J. Virol. 2013, 87, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Contesto-Richefeu, C.; Tarbouriech, N.; Brazzolotto, X.; Betzi, S.; Morelli, X.; Burmeister, W.P.; Iseni, F. Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminal domain. PLoS Pathog. 2014, 10, e1003978. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.A.; Stanitsa, E.S.; Greseth, M.D.; Lindgren, J.K.; Traktman, P. Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J. Biol. Chem. 2011, 286, 24702–24713. [Google Scholar] [CrossRef] [PubMed]
- Contesto-Richefeu, C.; Tarbouriech, N.; Brazzolotto, X.; Burmeister, W.P.; Peyrefitte, C.N.; Iseni, F. Structural analysis of point mutations at the vaccinia virus A20/D4 interface. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Tarbouriech, N.; Fender, P.; Contesto-Richefeu, C.; Peyrefitte, C.N.; Iseni, F. Crystal structure of the vaccinia virus uracil-DNA glycosylase in complex with DNA. J. Biol. Chem. 2015, 290, 17923–17934. [Google Scholar] [CrossRef] [PubMed]
- Schormann, N.; Sommers, C.I.; Prichard, M.N.; Keith, K.A.; Noah, J.W.; Nuth, M.; Ricciardi, R.P.; Chattopadhyay, D. Identification of protein-protein interaction inhibitors targeting vaccinia virus processivity factor for development of antiviral agents. Antimicrob. Agents Chemother. 2011, 55, 5054–5062. [Google Scholar] [CrossRef] [PubMed]
- Nuth, M.; Huang, L.; Saw, Y.L.; Schormann, N.; Chattopadhyay, D.; Ricciardi, R.P. Identification of inhibitors that block vaccinia virus infection by targeting the DNA synthesis processivity factor D4. J. Med. Chem. 2011, 54, 3260–3267. [Google Scholar] [CrossRef] [PubMed]
- Flusin, O.; Saccucci, L.; Contesto-Richefeu, C.; Hamdi, A.; Bardou, C.; Poyot, T.; Peinnequin, A.; Crance, J.M.; Colas, P.; Iseni, F. A small molecule screen in yeast identifies inhibitors targeting protein-protein interactions within the vaccinia virus replication complex. Antivir. Res. 2012, 96, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Tarbouriech, N.; Ducournau, C.; Hutin, S.; Mas, P.J.; Man, P.; Forest, E.; Hart, D.J.; Peyrefitte, C.N.; Burmeister, W.P.; Iseni, F. The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding. Nat. Commun. 2017, 8, 1455. [Google Scholar] [CrossRef] [PubMed]
- WHO. Report of the Eighteen Meeting; WHO Advisory Committee on Variola Virus Research: Geneva, Switzerland, 2016. [Google Scholar]
- Kupferschmidt, K. Labmade smallpox is possible, study shows. Science 2017, 357, 115–116. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delaune, D.; Iseni, F.; Ferrier-Rembert, A.; Peyrefitte, C.N.; Ferraris, O. The French Armed Forces Virology Unit: A Chronological Record of Ongoing Research on Orthopoxvirus. Viruses 2018, 10, 3. https://doi.org/10.3390/v10010003
Delaune D, Iseni F, Ferrier-Rembert A, Peyrefitte CN, Ferraris O. The French Armed Forces Virology Unit: A Chronological Record of Ongoing Research on Orthopoxvirus. Viruses. 2018; 10(1):3. https://doi.org/10.3390/v10010003
Chicago/Turabian StyleDelaune, Déborah, Frédéric Iseni, Audrey Ferrier-Rembert, Christophe N. Peyrefitte, and Olivier Ferraris. 2018. "The French Armed Forces Virology Unit: A Chronological Record of Ongoing Research on Orthopoxvirus" Viruses 10, no. 1: 3. https://doi.org/10.3390/v10010003



