1. Introduction
Human respiratory syncytial virus (HRSV) is the leading cause of acute lower respiratory tract infection in children less than 5 years of age, with high hospitalization rates in infants younger than 6 months [
1]. It also causes significant morbidity in adults, and contributes to excess mortality in older individuals and immunocompromised individuals of any age [
2,
3]. In the latter, infection may be prolonged and often complicated by bacterial co-infection, leading to pneumonia and severe respiratory distress [
4]. Outbreaks among vulnerable patients in hospital settings are of particular concern [
5,
6]. Nosocomial transmission poses a substantial risk and remains a serious challenge for hospital infection control. In the absence of an effective vaccine, HRSV treatment options are largely limited to supportive care and expensive monoclonal antibody or antiviral therapy [
7,
8]. Infection control measures, including isolation of cases, high standards of hand hygiene, cohort nursing, and use of personal protective equipment have been reported as relatively effective in the prevention and control of HRSV outbreaks in nosocomial settings [
5,
9,
10]. The prophylaxis use of monoclonal antibody therapy also can reduce nosocomial transmission [
11]. There is nonetheless a need for a larger armamentarium of HRSV-specific treatments and evidence-based control measures for HRSV infection prevention and control.
The lack of an appropriate animal model fully recapitulating HRSV pathogenesis of infection and disease has long represented a hurdle for the development of new intervention strategies against HRSV, including its spread in nosocomial settings [
12]. In humans, HRSV is transmitted by direct contact and large aerosol droplets, and via contaminated surfaces, initiating viral replication in the upper respiratory tract after an incubation period of 4–5 days [
2]. In severe cases, HRSV further spreads to and replicates in the lower respiratory tract. The virus replicates in ciliated epithelial cells along the respiratory tract, as well as in type I and type II alveolar pneumocytes, resulting in inflammation and necrosis. Animal models for HRSV infection are mostly semi-permissive, requiring a large inoculum of HRSV to result in a productive infection [
12]. They include non-human primate (NHP) models, which, apart from chimpanzees and African green monkeys, generally develop no or little clinical signs or pathological changes. The most used non-primate models of HRSV infection are cotton rats, mice, and the neonatal lamb. These models have contributed to improving our understanding of HRSV pathogenesis and host immune responses [
13,
14]. Furthermore, NHPs and cotton rats may also develop atypical enhanced disease following vaccination with formalin-inactivated HRSV, as seen in humans. However, they all have their limitations [
12]. In none of these models HRSV is transmitted to contact individuals, or they are highly impractical to study transmission.
The ferret model of HRSV infection had initially yielded disappointing results, with poor, age-dependent replication of the virus in the lower respiratory tract [
12,
15]. However, we reported recently that a low-passage clinical isolate of HRSV efficiently replicates in both the upper and lower respiratory tracts of adult ferrets [
16]. In addition, we described the use of immunocompromised ferrets as a relevant model for HRSV infection in immunocompromised individuals [
16]. It is characterized by high replication levels in the lower respiratory tract, with viral antigen detected in tracheal, bronchial, and bronchiolar ciliated epithelial cells, and by prolonged viral replication and shedding, reflecting the increased viral loads and delayed viral clearance observed in immunocompromised individuals. Prolonged infection is thought to be an important risk factor for nosocomial transmission [
17].
Prince et al. reported that infant ferrets generate higher virus loads than adult ferrets, resulting in successful contact transmission of HRSV from infant ferrets to their mother [
15]. Recently, Chan et al. showed limited horizontal transmission in a preliminary study between immunocompetent ferrets without proof of replicating virus in the contact ferrets [
18]. The immunocompromised ferret-HRSV model [
16] offers a new tool to study HRSV transmission and its prevention, which may be used to address the potential of intervention strategies, including vaccines and antibody or antiviral treatments, to block or limit HRSV transmission. Here we report the effective transmission of a low-passage clinical isolate of HRSV from immunocompromised ferrets to both immunocompetent and immunocompromised contact ferrets with the detection of replicating virus in both donor and contact ferrets. Both the intranasal and intratracheal route of infection were explored to determine which site of virus replication would be responsible for transmission of the virus.
2. Materials and Methods
2.1. Virus
A clinical isolate of HRSV subgroup A strain was isolated in human respiratory epithelium carcinoma (HEp-2) cells from a nasal lavage of an infant hospitalized at Erasmus Medical Centre, Rotterdam, The Netherlands, in 2011. The virus was passaged exclusively in HEp-2 cells, cultured as described previously [
16]. The challenge stock used in the animal experiments was passage 3, and was prepared as described previously [
16]. Virus titers were determined by end-point titration on monolayers of HEp-2 cells and cytopathic effect (CPE) screening six days after infection. Viral titers were calculated using the method of Spearman and Karber. The challenge stock tested negative for a large panel of respiratory human viruses and mycoplasma by RT-PCR.
2.2. Ferrets
Ferrets (
Mustela putorius furo) were 16- to 18-month-old purpose-bred males, which were seronegative for Aleutian disease virus, and RSV subgroup A and B. They were housed in groups of 2 to 12, as described previously [
16]. Male ferrets were selected irrespective of any HRSV-related parameter, because male ferrets would be more robust to endure the treatment procedures. The present experiment complied with all national and international guidelines for care and use of animals and was ethically approved on 15 December 2015 by the Dutch Central Committee Animal Experiments (CCD), protocol number AVD277002015161.
Immunocompromised ferrets received orally twice daily the immunosuppressive medication of 20 mg/kg of body weight (bw) Mycophenolate mofetil (MMF) (CellCept
®, Roche, Woerden, The Netherlands), 0.5 mg/kg bw tacrolimus (Prograft
®, Astellas Pharma BV, Leiderdorp, The Netherlands), 8 mg/kg bw prednisolone (Hospital Pharmacy, UMCN St Radboud, Nijmegen, The Netherlands), together with antibiotic prophylaxis of 10 mg/kg bw amoxicillin and 2.5 mg/kg bw clavulanic acid (Pharmachemie BV, Haarlem, The Netherlands), as described previously [
16,
19].
2.3. HRSV Transmission Experiment
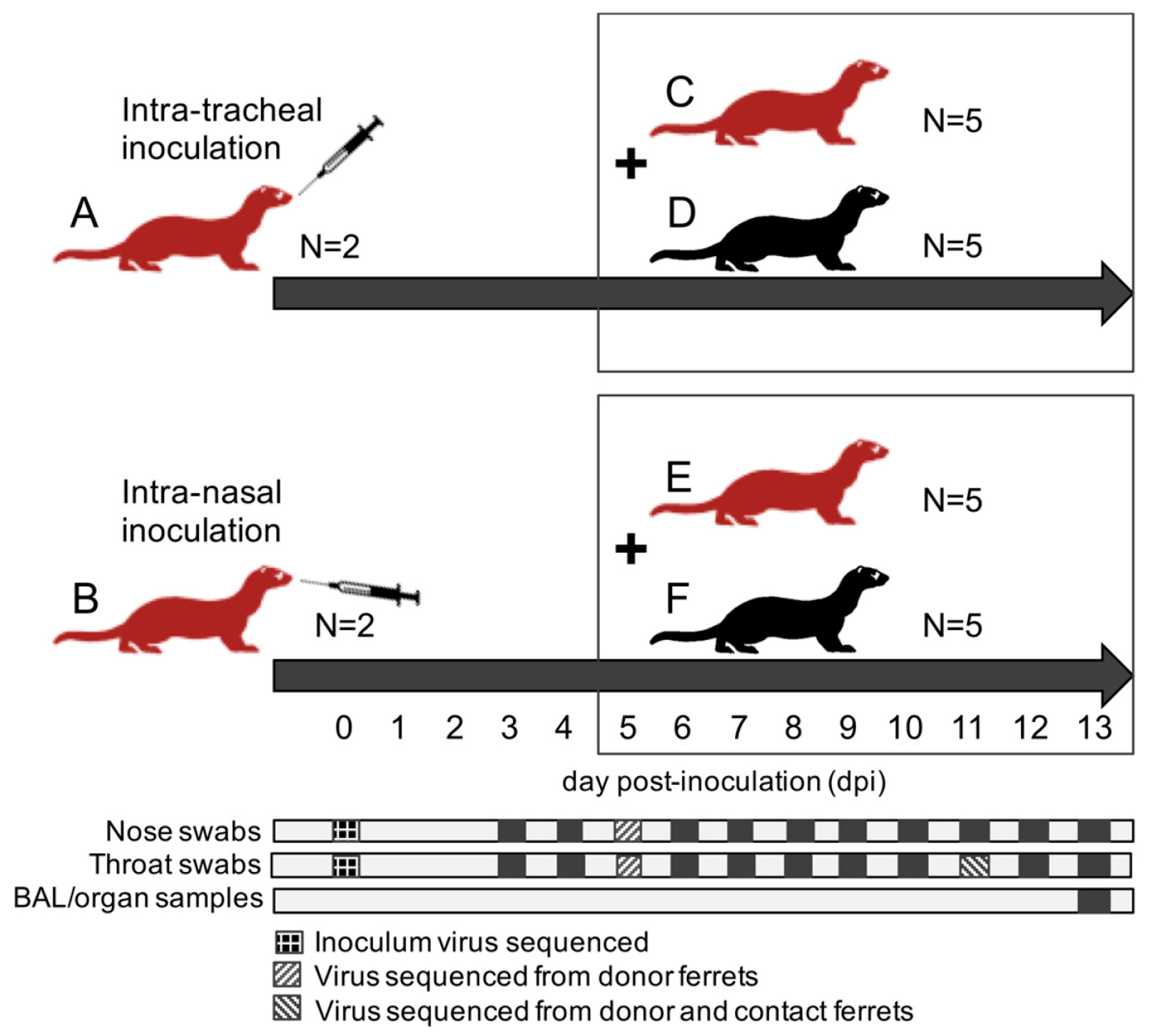
A total of twenty-four ferrets were randomly assigned to the following treatment groups (
Figure 1): (A) 2 immunocompromised donor ferrets infected with HRSV via the intra-tracheal route of inoculation; (B) 2 immunocompromised donor ferrets infected with HRSV via the intra-nasal route of inoculation; (C) 5 immunocompromised contact ferrets placed on day 5 post-inoculation (dpi) with group A ferrets; (D) 5 immunocompetent contact ferrets placed on 5 dpi with group A ferrets; (E) 5 immunocompromised contact ferrets placed on 5 dpi with group B ferrets; (F) 5 immunocompetent contact ferrets placed on 5 dpi with group B ferrets.
After a one-week acclimatization period, the ferrets assigned to group A, B, C, and E (
Figure 1) were started on the immunosuppression protocol, with daily oral prophylactic antibiotics starting four days before HRSV infection, and twice-daily oral immunosuppressants starting three days before HRSV infection. The ferrets assigned to group A and B were inoculated on day 0. Donor ferrets sedated with a mixture of ketamine and medetomidine (IM doses of 0.2 and 0.01 mL/kg body weight, respectively) were inoculated with 10
5 median tissue-culture infective dose (TCID
50) of low-passage clinical isolate HRSV subgroup A by intra-tracheal (IT; group A) or intra-nasal (IN; group B) inoculation with a volume of 3 or 0.3 mL, respectively.
The ferrets assigned to group C to F were introduced in the cages of group A and B ferrets on 5 dpi to enable direct contact. Throat (Copan; rayon tipped) and nose (Copan; polyester tipped) swabs were collected from all ferrets daily from 3 dpi onwards in a 3mL virus transport medium [
16]. Donor ferrets were euthanized by exsanguination under anesthesia with ketamine-medetomidine at 13 dpi and contact ferrets at day 8 post-exposure (dpe; corresponding to 13 dpi). Broncho-alveolar lavages (BAL) and samples of the nasal turbinates, trachea, primary bronchus, and lung parenchyma were collected upon autopsy.
All personnel involved in the collection of study data on a day-to-day basis and all personnel performing the laboratory analysis in which interpretation of the data is required were not aware of the so-called Random Treatment Allocation Key at any time prior to completion of the study. All samples were labeled with a unique sample number.
2.4. Samples and Assays
All samples were processed within four hours of sample collection. Infectious virus titers and concentrations of viral RNA were measured by virus isolation and reverse transcription-PCR (RT-PCR), respectively, as previously described [
16]. Samples of the nasal turbinates, trachea, primary bronchus, and right lung were weighed and subsequently homogenized with a FastPrep-24 (MP Biomedicals, Eindhoven, The Netherlands) in medium and centrifuged briefly before viral load assessment by virus isolation and quantitative RT-PCR.
RNA extraction was performed using a MagNA Pure 96 (Roche Diagnostics Nederland B.V., Almere, The Netherlands) and MagNA Pure 96 DNA and Viral NA Small Volume Kit with an input volume of 190 µL and output volume of 100 µL according to the manufacturer’s instructions (Roche Diagnostics Nederland B.V.). The extraction was internally controlled by the addition of a known concentration of phocine distemper virus (PDV) [
20]. 20 µL extracted RNA was amplified in a 50 µL final volume, containing 12.5 µL 4× TaqMan Fast Virus 1-Step Master Mix (Life Technologies, Bleiswijk, The Netherlands), and 1 µL of primers and probe mixture for detection of RSV A, RSV B, and PDV in a triplex reaction (
Table S1). The RT-PCR temperature profile was 5 min at 50 °C, 20 sec at 95 °C, 45 cycles of 3 s at 95 °C, and 32 s at 60 °C.
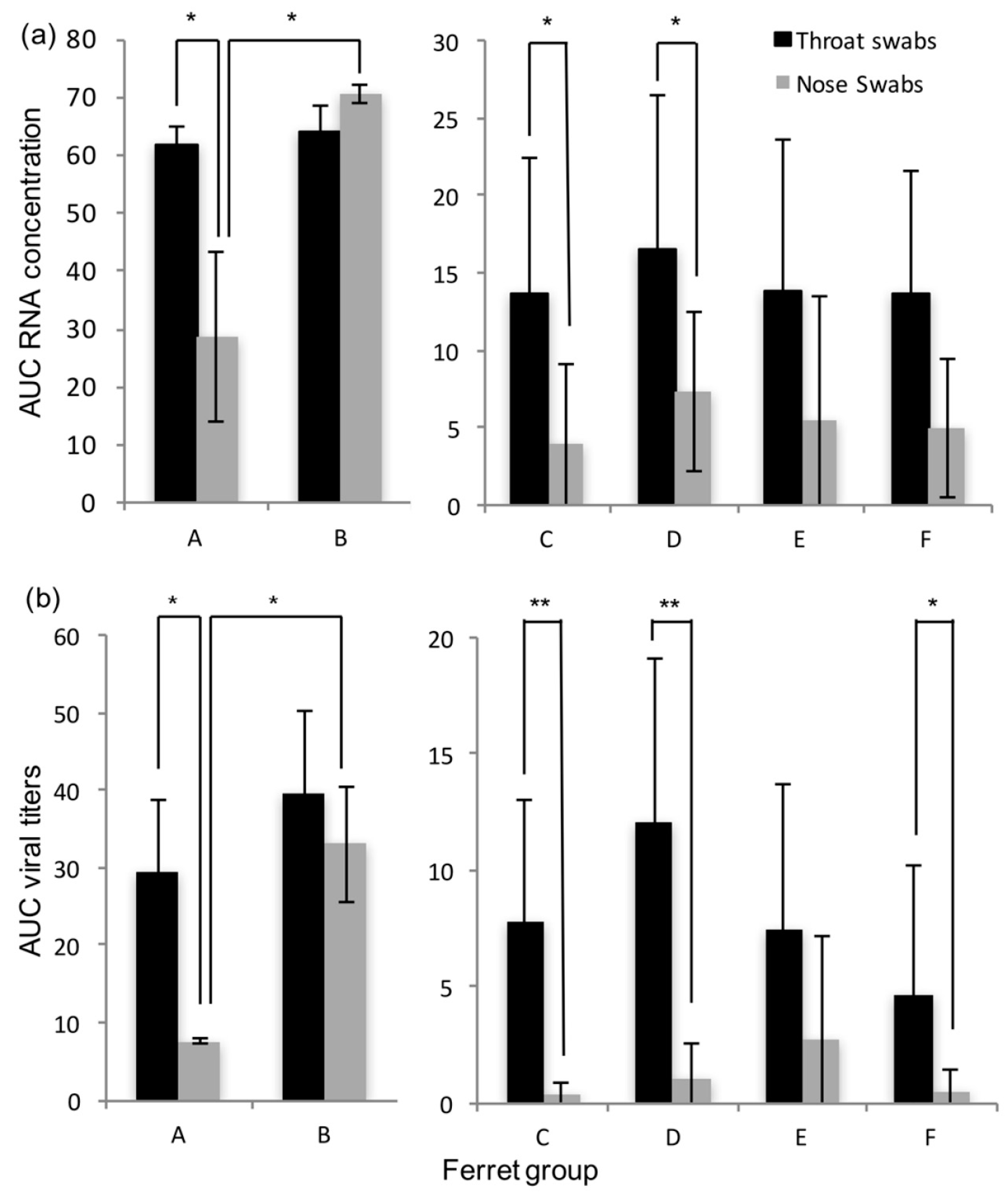
Dilutions of an electron microscopic-counted HRSV stock (ABIOnline.com Columbia) were used for conversion of RT-PCR threshold cycle (Ct) values into a quantitative measurement of viral particles. Concentrations of viral RNA are expressed as log10 vp/mL. Infectious virus titers in tissue are expressed as log10 TCID50 per gram tissue, and infectious virus titer in throat and nose swabs are expressed as log10 TCID50/mL. Significant differences in shedding loads were assessed based on the areas under the curve and using the Student t-test.
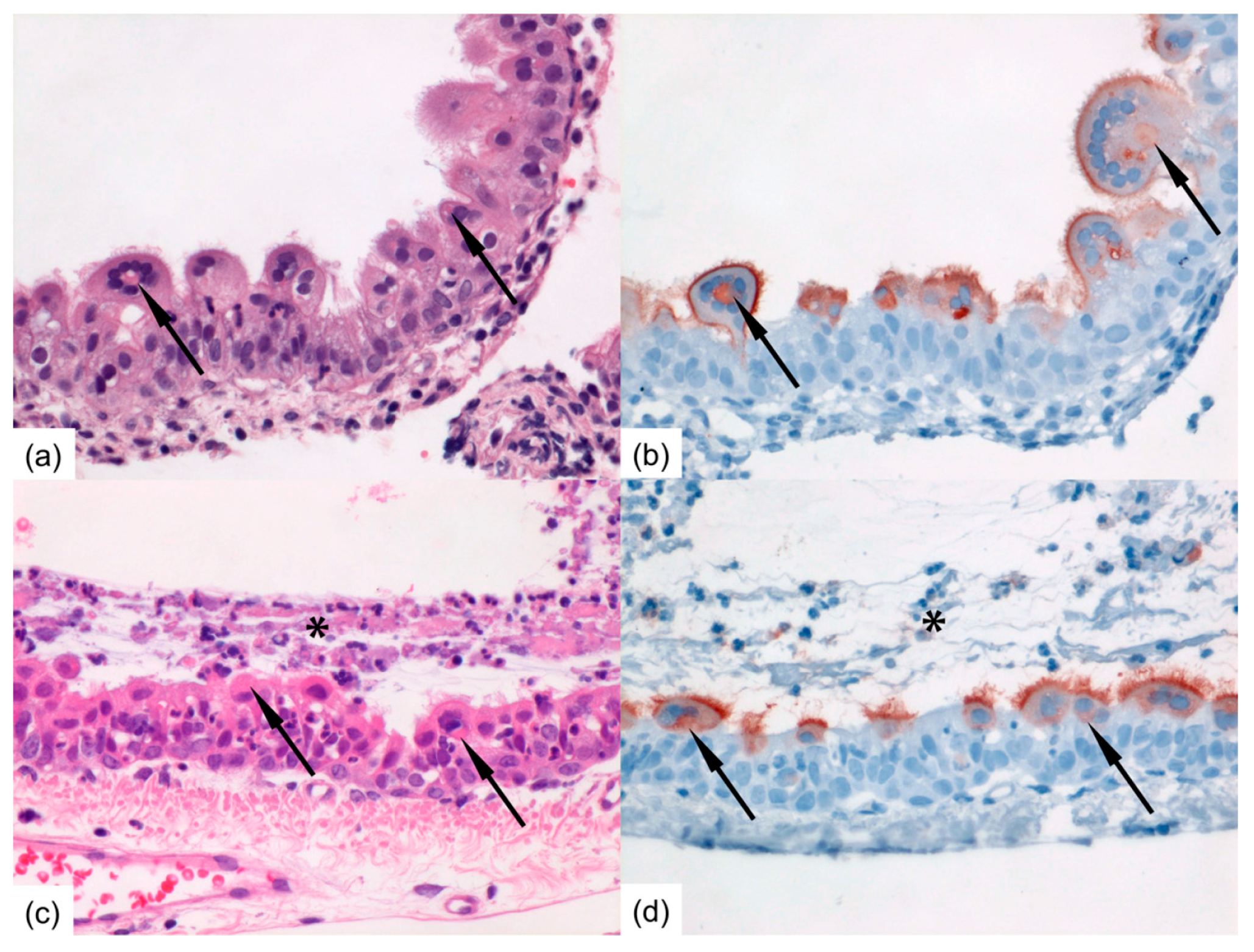
Formalin-fixed tissue sections of nasal turbinates, trachea, and left lung were routinely processed, paraffin embedded, and sectioned at 3–4 μm, deparaffinized with xylene and rehydrated using graded alcohols, and stained with hematoxylin and eosin (H&E) for histopathological examination by light microscopy. For immunohistochemistry (IHC), additional serial slides were sectioned simultaneously and incubated for 1 h with a goat anti-HRSV-peroxidase (PO) (Virostat, Portland, ME, USA) polyclonal antibody following antigen retrieval using citric acid buffer. Endogenous PO was blocked with 3% hydrogen peroxide. The bound PO was visualized by incubating slides with 3-amino-9-ethylcarbazole for 10 min as substrate, resulting in a reddish brown finely-granular staining of HRSV-infected epithelial cells, followed by hematoxylin counterstain. Negative controls were performed in the absence of the antibody.
2.5. Nested PCR Amplification and Sanger Sequencing
Full-length F and G gene sequencing was performed on the inoculum virus and RT-PCR positive nose and/or throat swabs collected from the donor ferrets at 5 and 11 dpi and from the contact ferrets at 6 dpe (corresponding to 11 dpi). RNA extraction was performed using a MagNA Pure 96 (Roche Diagnostics) and MagNA Pure 96 DNA and Viral NA Small Volume Kit with an input volume of 200 µL and output volume of 100 µL according to the manufacturer’s instructions (Roche Applied Science). cDNA synthesis was performed using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Semi-nested PCR amplification was performed using HotStarTaq DNA Polymerase (Qiagen, Hilden, Germany). Sequence reactions were performed with the BigDye
®Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI Prism 3730 genetic analyzer. Primers are indicated in
Table S1.
4. Discussion
Immunocompromised ferrets inoculated with HRSV can transmit the virus to contact ferrets that are either immunocompromised or immunocompetent. In general, HRSV viral loads were found similar in immunocompromised and immunocompetent contact ferrets. Although immunocompromised ferrets inoculated intra-nasally tended to have higher RNA or viral loads in the nose swabs than immunocompromised ferrets inoculated intra-tracheally, HRSV was transmitted as efficiently from either group to contact ferrets.
The transmission rate of HRSV in our ferret model using a low-passage HRSV isolate is high, reaching 80 to 100%, based on RT-PCR results. In contrast to the study by Chan et al. [
18], we chose to use the immunocompromised ferret model as donors for transmission studies, as it is more relevant for the clinical situation, and in addition more and longer HRSV shedding was observed [
16]. It demonstrates the potential of these models to be used for the quantitative assessment of HRSV transmission in the context of intervention studies. The immunocompromised donor ferrets reproduced a prolonged infection with high RNA and viral loads in nose and throat swabs as described previously, confirming the robustness of the model [
16]. Ferrets inoculated intra-nasally developed higher RNA and viral loads in the nose than ferrets inoculated intra-tracheally. Ferrets inoculated intra-nasally also tended to have more cells positive for HRSV-antigen in the upper regions of the respiratory tract, i.e., in the nasal turbinates and trachea than ferrets inoculated intra-tracheally. Conversely, ferrets inoculated intra-tracheally tended to have more positive cells for HRSV-antigen deeper down the respiratory tract, in the bronchi and bronchioles, than ferrets inoculated intra-nasally. This supports the fact that the route of transmission can have an impact on HRSV kinetics upon infection, as previously shown for influenza A virus in ferrets [
21,
22].
Both immunocompromised and immunocompetent contact ferrets effectively developed a productive infection following contact transmission of HRSV. In both donor and contact immunocompromised and immunocompetent ferrets, ciliated epithelial cells along the respiratory tract, from the nasal turbinates to the bronchi and bronchioles, were found infected, in association with inflammatory lesions. Only a few substitutions were sporadically detected in the F and G proteins of HRSV shed by contact ferrets, suggesting that little adaptation to the ferret model had occurred during the prolonged infection in donor immunocompromised ferrets and upon contact transmission. These observations further support the appropriateness of the ferret model to reproduce HRSV infection and pathogenesis as seen in humans, including after contact transmission of the virus. The use of a contact transmission model nonetheless precludes the assessment of the respective role of large droplets and aerosols in the transmission of HRSV. However, and despite differences in RNA and viral loads in the nose, donor ferrets inoculated intra-nasally and donor ferrets inoculated intra-tracheally transmitted HRSV to contact ferrets with similar efficiency. This suggests that HRSV transmission in these ferret models does not correlate with viral loads in the upper respiratory tract. Future studies using molecularly cloned HRSV constructs expressing a reporter gene, like EGFP, may facilitate studies of transmission mechanisms of HRSV.
In conclusion, the ferret models presented in this paper can be used to mimic nosocomial transmission of HRSV in vulnerable patients, as well as population-based transmission in humans, and also can be used to evaluate intervention strategies for the prevention of HRSV transmission.