Viroid Replication: Rolling-Circles, Enzymes and Ribozymes
Abstract
:1. Introduction
2. An overview of viroid singularity and diversity
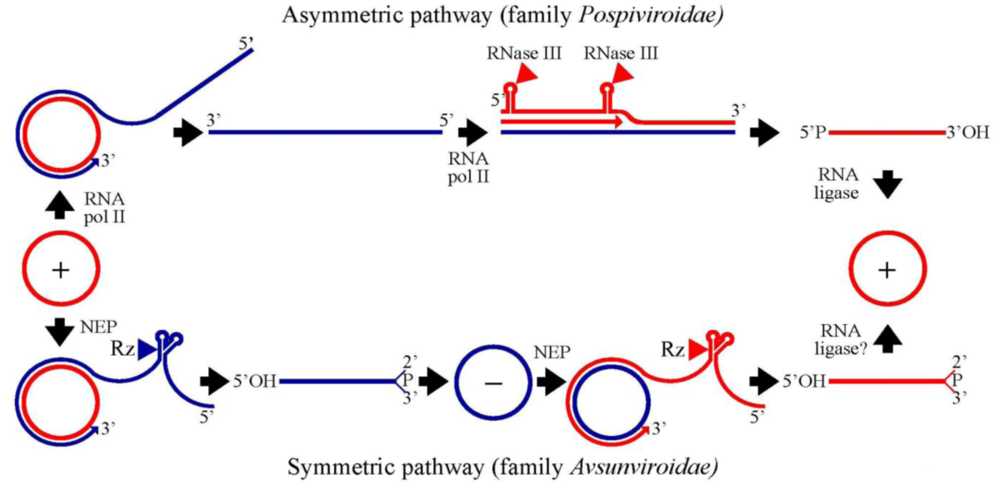
3. Family Pospiviroidae: an asymmetric rolling-circle mechanism with circular and linear templates catalyzed by host enzymes
3.1. Initiation and elongation: involvement of RNA polymerase II
3.2. Cleavage of oligomeric (+) strands is most likely mediated by a member of the RNase III family
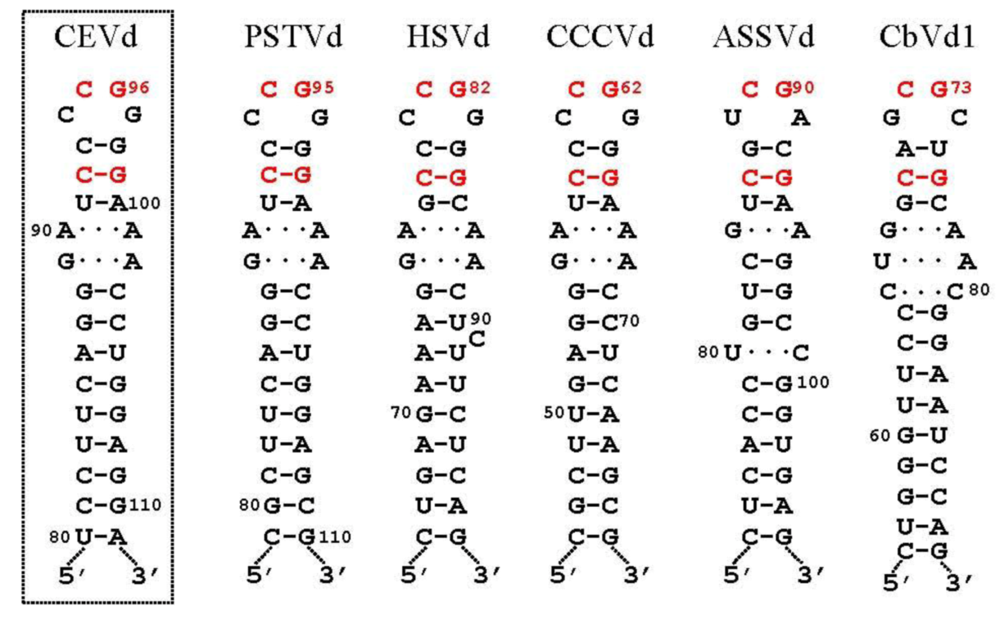
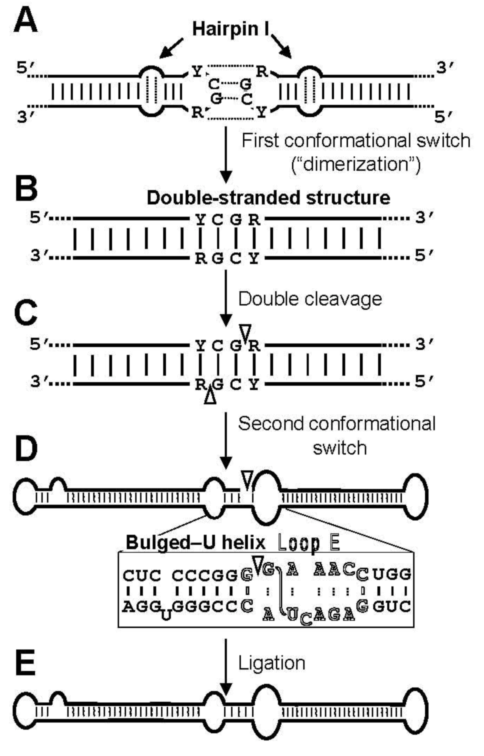
3.3. Ligation of monomeric linear (+) strands: presumable participation of a novel RNA ligase
4. Family Avsunviroidae: a symmetric rolling-circle mechanism with circular RNA templates catalyzed by host enzymes and viroid ribozymes
4.1. Initiation and elongation: involvement of a nuclear-encoded chloroplastic RNA polymerase
4.2. Cleavage of oligomeric (+) and (-) strands is mediated by hammerhead ribozymes
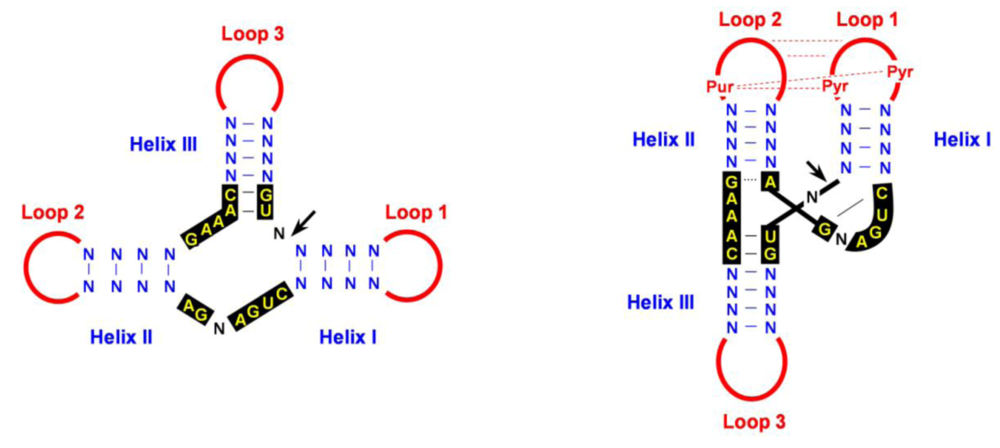
4.3. Ligation: an unsolved question with several possible alternatives
5. Concluding remarks and perspectives
Acknowledgments
References
- Gross, H.J.; Domdey, H.; Lossow, C.; Jank, P.; Raba, M.; Alberty, H.; Sänger, H.L. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978, 273, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 2003, 1, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Tabler, M.; Tsagris, M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004, 9, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Hernández, C.; Martínez de Alba, E.; Daròs, J.A.; Di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.A.; Elena, S.F.; Flores, R. Viroids: an Ariadne’s thread into the RNA labyrinth. EMBO Rep. 2006, 7, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Itaya, A. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microb. Interact. 2007, 20, 7–20. [Google Scholar] [CrossRef]
- Ding, B. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef] [PubMed]
- Astier-Manifacier, S.; Cornuet, P. RNA-dependent RNA polymerase in Chinese cabbage. Biochim. Biophys. Acta 1971, 232, 484–493. [Google Scholar] [PubMed]
- Schiebel, W.; Haas, B.; Marinkovic, S.; Klanner, A.; Sanger, H. L. RNA-directed RNA polymerase from tomato leaves. 1. Purification and physical-properties. J. Biol. Chem. 1993, 268, 11851–11857. [Google Scholar] [PubMed]
- Przybilski, R.; Graf, S.; Lescoute, A.; Nellen, W.; Westhof, E.; Steger, G.; Hammann, C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 2005, 17, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Martick, M.; Horan, L.H.; Noller, H.F.; Scott, W.G. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 2008, 454, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Daròs, J.A.; Hernández, C. The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res. 2000, 55, 271–323. [Google Scholar] [PubMed]
- Symons, R.H.; Randles, J.W. Encapsidated circular viroid-like satellite RNAs (virusoids) of plants. Curr. Top. Microbiol. Immunol. 1999, 239, 81–105. [Google Scholar] [PubMed]
- Lai, M.M.C. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 2005, 79, 7951–7958. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.D.; Robertson, H.D. A replication cycle for viroids and other small infectious RNAs. Science 1984, 223, 450–454. [Google Scholar] [PubMed]
- Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 1988, 85, 9128–9132. [CrossRef]
- Zhong, X.; Archual, A.J.; Amin, A.A.; Ding, B. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 2008, 20, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Kolonko, N.; Bannach, O.; Aschermann, K.; Hu, K.H.; Moors, M.; Schmitz, M.; Steger, G.; Riesner, D. Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 2006, 347, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Riesner, D.; Henco, K.; Rokohl, U.; Klotz, G.; Kleinschmidt, A.K.; Domdey, H.; Jank, P.; Gross, H.J.; Sänger, H.L. Structure and structure formation of viroids. J. Mol. Biol. 1979, 133, 85–115. [Google Scholar] [CrossRef] [PubMed]
- Tabler, M.; Sänger, H.L. Infectivity studies on different potato spindle tuber viroid (PSTV) RNAs synthesized in vitro with the SP6 transcription system. EMBO J. 1985, 4, 2191–2199. [Google Scholar] [PubMed]
- Meshi, T.; Ishikawa, M; Watanabe, Y.; Yamaya, J.; Okada, Y.; Shikata, E. The sequence necessary for the infectivity of hop stunt viroid cDNA clones. Mol. Gen. Genet. 1985, 200, 199–206. [Google Scholar] [CrossRef]
- Visvader, J.E.; Forster, A.C.; Symons, R.H. Infectivity and in vitro mutagenesis of monomeric cDNA clones of citrus exocortis viroid indicates the site of processing of viroid precursors. Nucleic Acids Res. 1985, 13, 5843–5856. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Viroid processing: a model involving the central conserved region and hairpin I. Proc. Natl. Acad. Sci. USA 1986, 83, 58–62. [Google Scholar] [CrossRef]
- Baumstark, T.; Schröder, A.R.; Riesner, D. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 1997, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Di Serio, F.; Hernández, C. Viroids: the noncoding genomes. Semin. Virol. 1997, 8, 65–73. [Google Scholar] [CrossRef]
- Gas, M.E.; Hernández, C.; Flores, R.; Daròs, J.A. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 2007, 3, 1813–1826. [Google Scholar] [CrossRef]
- Daròs, J.A.; Flores, R. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. USA 2004, 101, 6792–6797. [Google Scholar] [CrossRef]
- Gast, F.U.; Kempe, D.; Sänger, H.L. The dimerization domain of potato spindle tuber viroid, a possible hallmark for infectious RNA. Biochemistry 1998, 37, 14098–14107. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- MacRae, I.J.; Doudna, J.A. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007, 17, 1–8. [Google Scholar] [CrossRef]
- Hiraguri, A.; Itoh, R.; Kondo, N.; Nomura, Y.; Aizawa, D.; Murai, Y.; Koiwa, H.; Seki, M.; Shinozaki, K.; Fukuhara, T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Gas, M.E.; Molina-Serrano, D.; Hernández, C.; Flores, R.; Daròs, J.A. Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5’-phosphomonoester and 3’-hydroxyl termini: implications for the ribonuclease and RNA ligase involved in replication. J. Virol. 2008, 82, 10321–10325. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Shehu-Xhilaga, M.; Marquet, R.; Mak, J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004, 2, 461–472. [Google Scholar] [CrossRef]
- Branch, A.D.; Robertson, H.D.; Greer, C.; Gegenheimer, P.; Peebles, C.; Abelson, J. Cell-free circularization of viroid progeny RNA by an RNA ligase from wheat germ. Science 1982, 217, 1147–1149. [Google Scholar] [PubMed]
- Konarska, M.; Filipowicz, W.; Domdey, H.; Gross, H.J. Formation of a 2’-phosphomonoester, 3’,5’-phosphodiester linkage by a novel RNA ligase in wheat germ. Nature 1981, 293, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Tabler, M.; Tzortzakaki, S.; Tsagris, M. Processing of linear longer-than-unit-length potato spindle tuber viroid RNAs into infectious monomeric circular molecules by a G-specific endoribonuclease. Virology 1992, 190, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.D.; Benenfeld, B.J.; Robertson, H.D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. USA 1985, 82, 6590–6594. [Google Scholar] [CrossRef]
- Eiras, M.; Kitajima, E.W.; Flores, R.; Daròs, J.A. Existence in vivo of the loop E motif in potato spindle tuber viroid RNA. Arch. Virol. 2007, 152, 1389–1393. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, X.; Itaya, A.; Ding, B. Evidence for the existence of the loop E motif of potato spindle tuber viroid in vivo. J. Virol. 2007, 81, 2074–2077. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M.; Spieker, R.L.; Thalmeir, S.; Gast, F.U.; Riedel, L.; Sänger, H.L. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 1996, 226, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ding, B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 2003, 15, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Leontis, N.; Qiang, S.; Itaya, A.; Qi, Y.; Boris-Lawrie, K.; Ding, B. Tertiary structural and functional analysis of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J. Virol. 2006, 80, 8566–8581. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.K.; Shuman, S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. USA 2002, 99, 12709–12714. [Google Scholar] [CrossRef]
- Wang, L.K.; Schwer, B.; Shuman, S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA 2006, 12, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, C.; Rathjen, P.D.; Forster, A.C; Symons, R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.A.; Marcos, J.F.; Hernández, C.; Flores, R. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 1994, 91, 12813–12817. [Google Scholar] [CrossRef]
- Hernández, C.; Flores, R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef]
- Navarro, B.; Flores, R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 1997, 94, 11262–11267. [Google Scholar] [CrossRef]
- Fadda, Z.; Daròs, J.A.; Fagoaga, C.; Flores, R.; Duran-Vila, N. Eggplant latent viroid (ELVd): candidate type species for a new genus within family Avsunviroidae (hammerhead viroids). J. Virol. 2003, 77, 6528–6532. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Hernández, C.; De la Peña, M.; Vera, A.; Daròs, J.A. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 2001, 341, 540–552. [Google Scholar] [PubMed]
- Navarro, J.A.; Vera, A.; Flores, R. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 2000, 268, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Rodio, M.E.; Delgado, S.; De Stradis, A.E.; Gómez, M.D.; Flores, R.; Di Serio, F. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 2007, 19, 3610–3626. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.A.; Flores, R. Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J. 2000, 19, 2662–2670. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; Martínez de Alba, E.; Hernández, C.; Flores, R. A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 2005, 79, 12934–12943. [Google Scholar] [CrossRef] [PubMed]
- Motard, J.; Bolduc, F.; Thompson, D.; Perreault, J.P. The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 2008, 373, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Pelchat, M.; Côté, F.; Perreault, J.P. Study of the polymerization step of the rolling circle replication of peach latent mosaic viroid. Arch. Virol. 2001, 146, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Circular RNAs: relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- De la Peña, M.; Gago, S.; Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003, 22, 5561–5570. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Lescoute, A.; Westhof, E.; Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003, 10, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Martick, M.; Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006, 126, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.I.; Martick, M.; Lares, M.; Kim, R.; Scott, W.G.; Kim, S.H. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008, 6, 2060–2068. [Google Scholar]
- Dufour, D.; De la Peña, M.; Gago, S.; Flores, R.; Gallego, J. Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009, 37, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.A.; Flores, R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002, 21, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Molina-Serrano, D.; Suay, L.; Salvador, M.L.; Flores, R.; Daròs, J.A. Processing of RNAs of the family Avsunviroidae in Chlamydomonas reinhardtii chloroplasts. J. Virol. 2007, 81, 4363–4366. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; De la Peña, M.; Flores, R.; Gago, S. Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: differences in co- and post-transcriptional self-cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res. 2006, 34, 5613–5622. [Google Scholar] [CrossRef] [PubMed]
- Englert, M; Beier, H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005, 33, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.W.; Abelson, J. The subnuclear localization of transfer-RNA ligase in yeast. J. Cell Biol. 1987, 105, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Yoshihisa, T; Yunoki-Esaki, K; Ohshima, C.; Tanaka, N.; Endo, T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell 2003, 14, 3266–3279. [Google Scholar] [CrossRef] [PubMed]
- Englert, M; Latz, A; Becker, D.; Gimple, O.; Beier, H.; Akama, K. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 2007, 89, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Kibertis, P.A.; Haseloff, J.; Zimmern, D. 2’ phosphomonoester, 3’-5’ phosphodiester bond at a unique site in a circular viral RNA. EMBO J. 1985, 4, 817–827. [Google Scholar] [PubMed]
- Ambrós, S.; Hernández, C.; Desvignes, J.C; Flores, R. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: Implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J. Virol. 1998, 72, 7397–7406. [Google Scholar] [PubMed]
- Ambrós, S.; Hernández, C.; Flores, R. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 1999, 80, 2239–2252. [Google Scholar] [PubMed]
- De la Peña, M.; Navarro, B.; Flores, R. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. USA 1999, 96, 9960–9965. [Google Scholar] [CrossRef]
- Buzayan, J.M.; Gerlach, W.L.; Bruening, G. Nonenzymatic cleavage and ligation of RNAs complementary to a plant-virus satellite RNA. Nature 1986, 323, 349–353. [Google Scholar] [CrossRef]
- Stage-Zimmermann, T.K.; Uhlenbeck, O.C. A covalent crosslink converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nat. Struct. Biol. 2001, 8, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.A.; Shepotinovskaya, I.; Uhlenbeck, O.C. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry 2005, 44, 14577–14585. [Google Scholar] [CrossRef] [PubMed]
- Côté, F.; Perreault, J.P. Peach latent mosaic viroid is locked by a 2’,5’- phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 1997, 273, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Côté, F.; Lévesque, D.; Perreault, J.P. Natural 2’,5’-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J. Virol. 2001, 75, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sharmeen, L.; Kuo, M.Y.P.; Taylor, J. Self-ligating RNA sequences on the antigenome of human hepatitis delta-virus. J. Virol. 1989, 63, 1428–1430. [Google Scholar] [PubMed]
- Reid, C.E.; Lazinski, D.W. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 2000, 97, 424–429. [Google Scholar] [CrossRef]
- Marcos, J.F.; Flores, R. The 5' end generated in the in vitro self-cleavage reaction of avocado sunblotch viroid RNAs is present in naturally occurring linear viroid molecules. J. Gen. Virol. 1993, 74, 907–910. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.B.; Bian, X.Y.; Wu, L.M.; Liu, L.X.; Smith, N.A.; Isenegger, D.; Wu, R.M.; Masuta, C.; Vance, V.B.; Watson, J.M.; Rezaian, A.; Dennis, E.S.; Waterhouse, P.M. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; Wang, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Gago, S.; Elena, S.F.; Flores, R.; Sanjuán, R. Extremely high variability of a hammerhead viroid. Science 2009, 323, 1308. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Flores, R.; Gas, M.-E.; Molina-Serrano, D.; Nohales, M.-Á.; Carbonell, A.; Gago, S.; De la Peña, M.; Daròs, J.-A. Viroid Replication: Rolling-Circles, Enzymes and Ribozymes. Viruses 2009, 1, 317-334. https://doi.org/10.3390/v1020317
Flores R, Gas M-E, Molina-Serrano D, Nohales M-Á, Carbonell A, Gago S, De la Peña M, Daròs J-A. Viroid Replication: Rolling-Circles, Enzymes and Ribozymes. Viruses. 2009; 1(2):317-334. https://doi.org/10.3390/v1020317
Chicago/Turabian StyleFlores, Ricardo, María-Eugenia Gas, Diego Molina-Serrano, María-Ángeles Nohales, Alberto Carbonell, Selma Gago, Marcos De la Peña, and José-Antonio Daròs. 2009. "Viroid Replication: Rolling-Circles, Enzymes and Ribozymes" Viruses 1, no. 2: 317-334. https://doi.org/10.3390/v1020317




