Estimating Stand Density in a Tropical Broadleaf Forest Using Airborne LiDAR Data
Abstract
:1. Introduction
2. Materials
2.1. Study Area
2.2. ALS Dataset
2.3. In-Situ Measurements
3. Methods
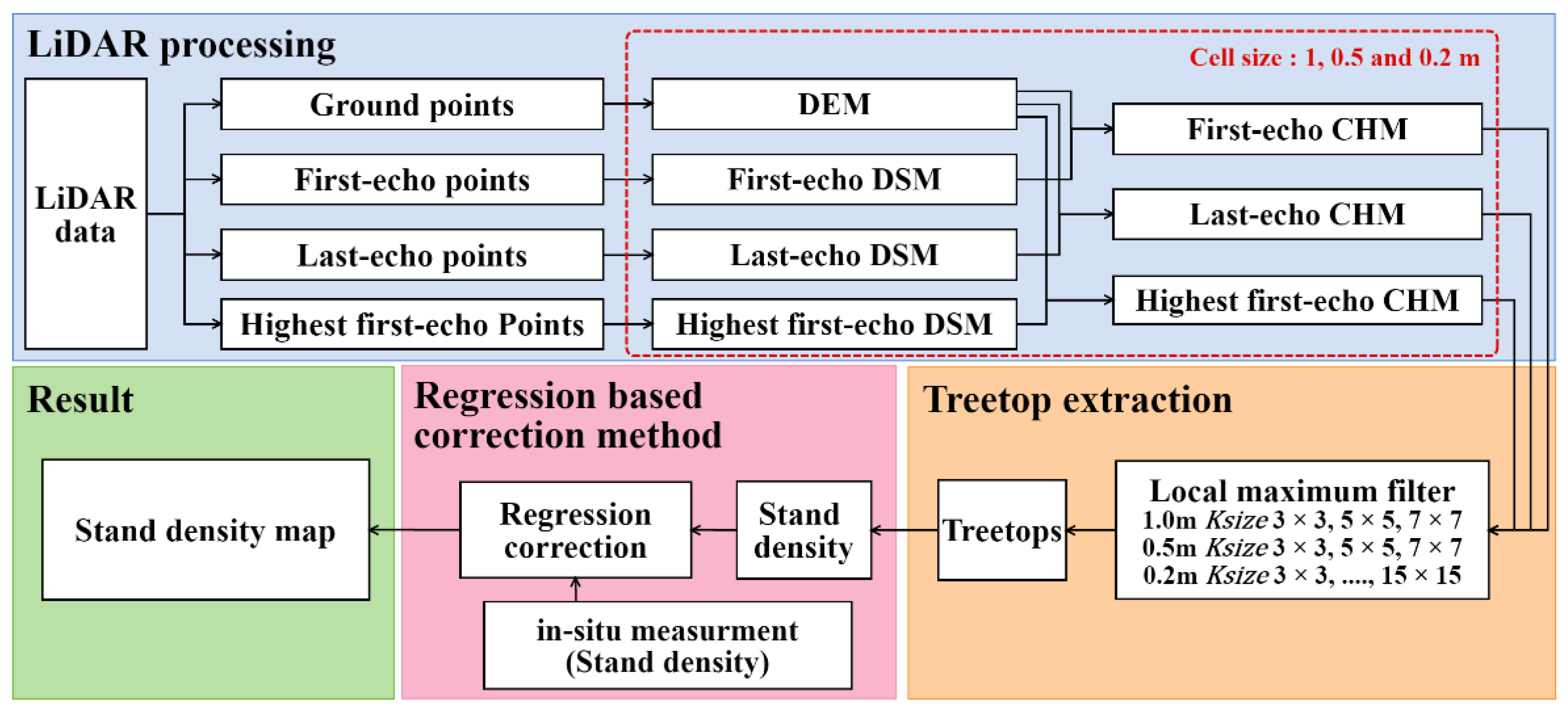
3.1. Steps of Stand Density Estimation
3.2. DSM, DEM and CHM Generation
3.3. Stand Density Estimaion From Treetop Extraction Using the Local Maximum Method
3.4. Regression-Based Correction Method
3.5. Error Assessment
3.6. Leave-One-Out Cross-Validation
3.7. Numerical LiDAR Data Thinning
4. Results
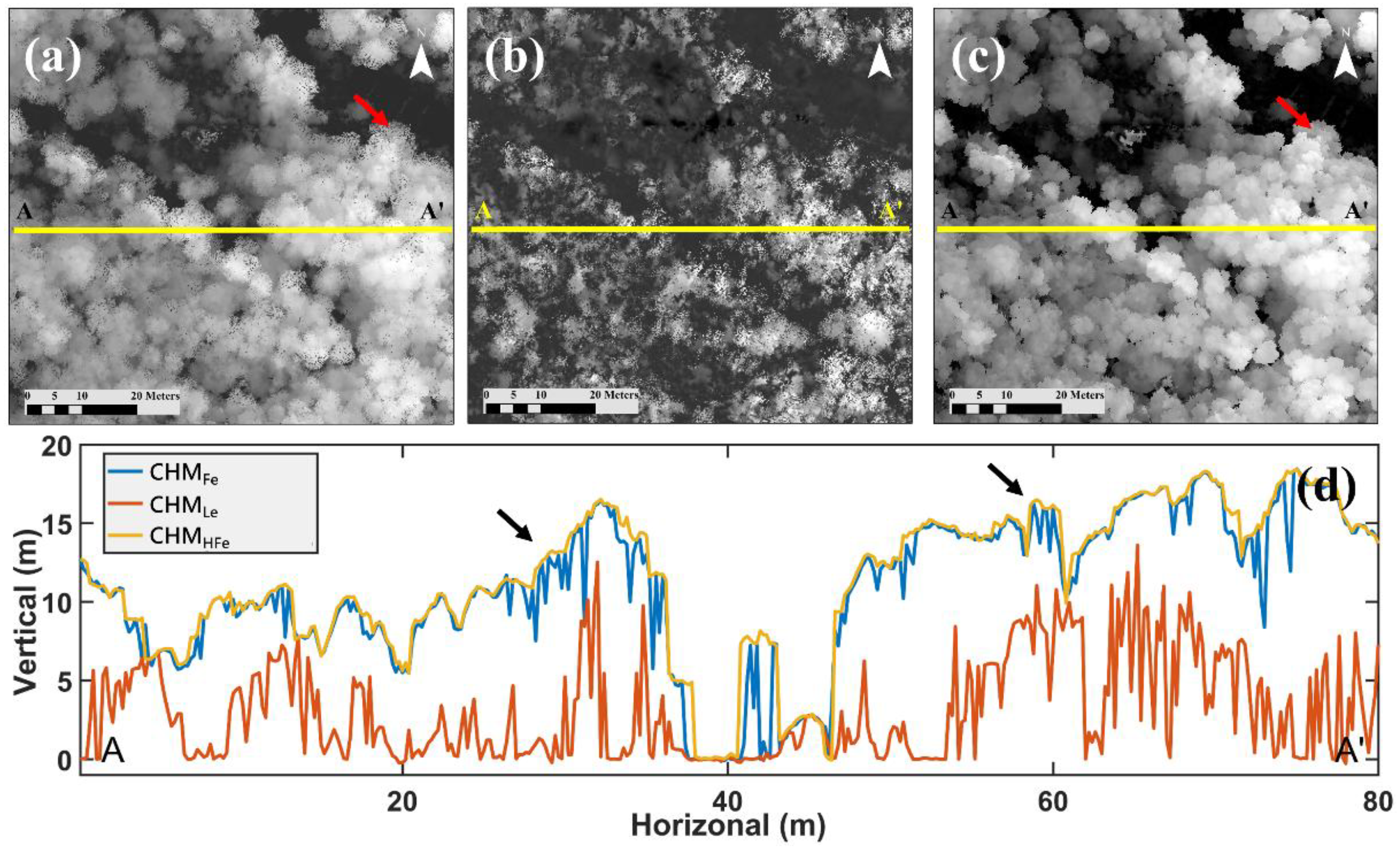
4.1. Comparison of Three Types CHM Data
4.2. Stand Density by the Local Maximum Method
4.3. Using Leave-One-Out Cross-Validation to Test Stand Density Errors
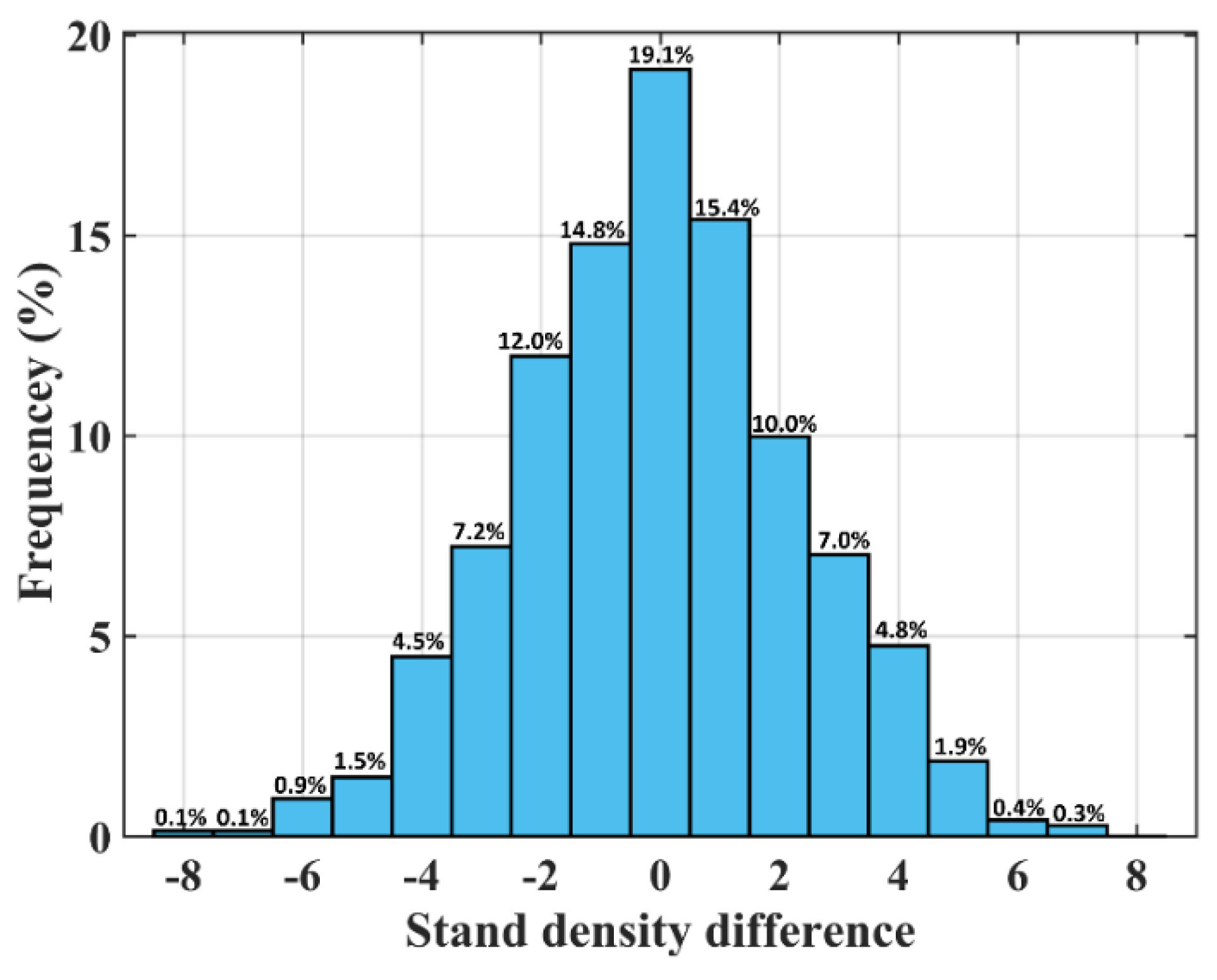
4.4. Stand Density Map by the Regression-Based Correction Method
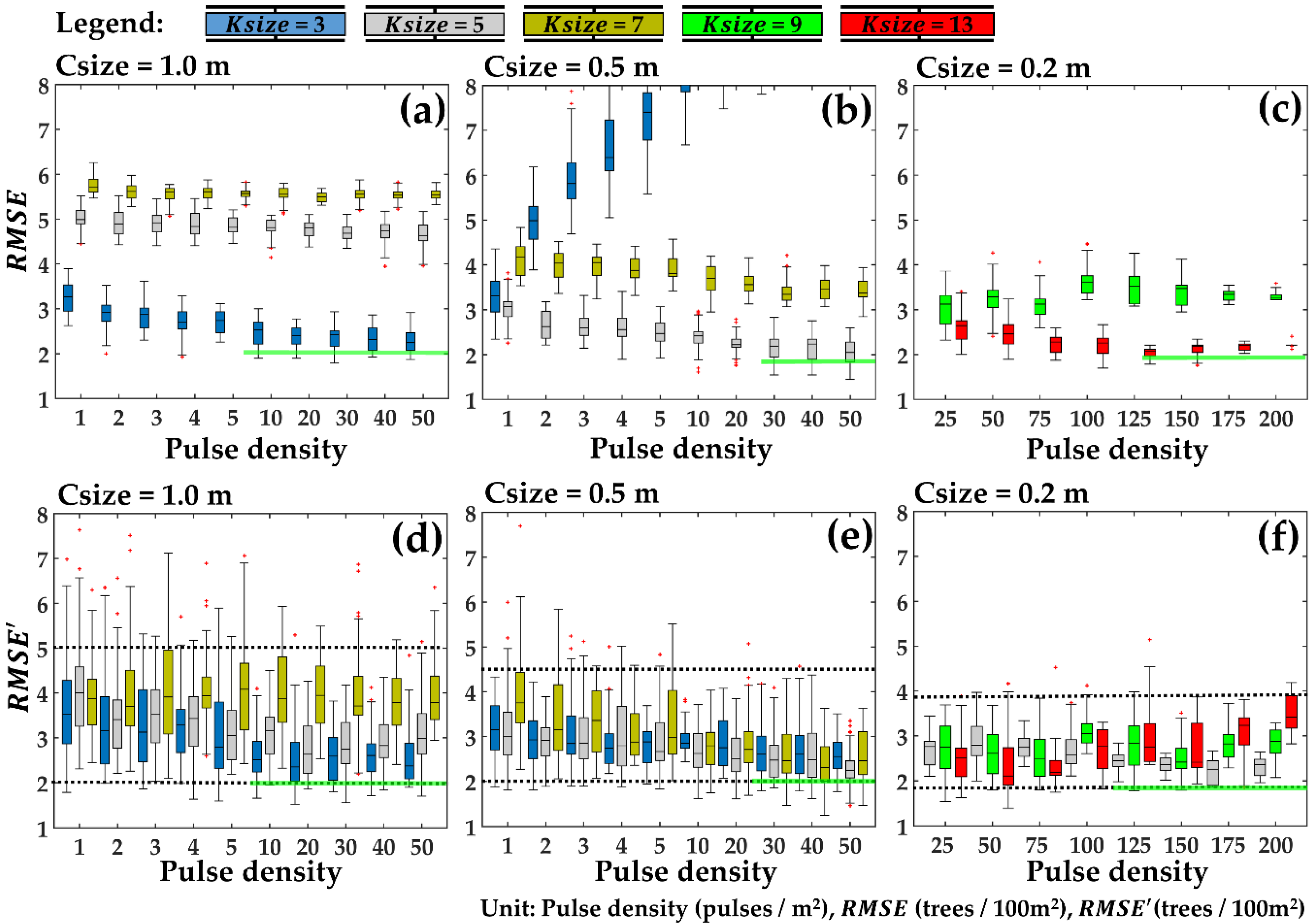
4.5. Error Assessment of Stand Density Estimation at Different Laser Pulse Densities
5. Discussions
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lorimer, C.G.; White, A.S. Scale and frequency of natural disturbances in the northeastern US: Implications for early successional forest habitats and regional age distributions. For. Ecol. Manag. 2003, 185, 41–64. [Google Scholar] [CrossRef]
- Espirito-Santo, F.D.B.; Gloor, M.; Keller, M.; Malhi, Y.; Saatchi, S.; Nelson, B.; Oliveira, R.C.; Pereira, C.; Lloyd, J.; Frolking, S.; et al. Size and frequency of natural forest disturbances and the Amazon forest carbon balance. Nat. Commun. 2014, 5, 3434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeide, B. How to measure stand density. TreesStruct. Funct. 2005, 19, 1–14. [Google Scholar] [CrossRef]
- Keddy, P.A. Plants and Vegetation: Origins, Processes, Consequences; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2007; p. 683. [Google Scholar]
- Ulvcrona, K.A.; Claesson, S.; Sahlen, K.; Lundmark, T. The effects of timing of pre-commercial thinning and stand density on stem form and branch characteristics of Pinus sylvestris. Forestry 2007, 80, 323–335. [Google Scholar] [CrossRef]
- Franklin, O.; Aoki, K.; Seidl, R. A generic model of thinning and stand density effects on forest growth, mortality and net increment. Ann. For. Sci. 2009, 66, 815. [Google Scholar] [CrossRef]
- Wu, Y.C.; Strahler, A.H. Remote estimation of crown size, stand density, and biomass on the Oregon transect. Ecol. Appl. 1994, 4, 299–312. [Google Scholar] [CrossRef]
- Sousa, V.B.; Louzada, J.L.; Pereira, H. Variation of ring width and wood density in two unmanaged stands of the Mediterranean Oak Quercus faginea. Forests 2018, 9, 44. [Google Scholar] [CrossRef]
- Cheng, X.R.; Yu, M.K.; Wu, T.G. Effect of forest structural change on carbon storage in a coastal metasequoia glyptostroboides stand. Sci. World J. 2013, 2013, 830509. [Google Scholar] [CrossRef] [PubMed]
- Uhl, E.; Biber, P.; Ulbricht, M.; Heym, M.; Horváth, T.; Lakatos, F.; Gál, J.; Steinacker, L.; Tonon, G.; Ventura, M.; et al. Analysing the effect of stand density and site conditions on structure and growth of oak species using Nelder trials along an environmental gradient: Experimental design, evaluation methods, and results. For. Ecosyst. 2015, 2, 17. [Google Scholar] [CrossRef]
- Moon, M.; Kim, T.; Park, J.; Cho, S.; Ryu, D.; Kim, H.S. Variation in sap flux density and its effect on stand transpiration estimates of Korean pine stands. J. For. Res. 2015, 20, 85–93. [Google Scholar] [CrossRef]
- Humagain, K.; Portillo-Quintero, C.; Cox, R.D.; Cain, J.W. Mapping tree density in forests of the southwestern USA using Landsat 8 data. Forests 2017, 8, 287. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Naesset, E.; Orka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. LiDAR sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.F. Estimating plot-level tree heights with LiDAR: Local filtering with a canopy-height based variable window size. Comput. Electron. Agric. 2002, 37, 71–95. [Google Scholar] [CrossRef]
- Kwak, D.A.; Lee, W.K.; Lee, J.H.; Biging, G.S.; Gong, P. Detection of individual trees and estimation of tree height using LiDAR data. J. For. Res. 2007, 12, 425–434. [Google Scholar] [CrossRef]
- Barnes, C.; Balzter, H.; Barrett, K.; Eddy, J.; Milner, S.; Suarez, J.C. Individual tree crown delineation from airborne laser scanning for diseased larch forest stands. Remote Sens. 2017, 9, 231. [Google Scholar] [CrossRef]
- Gougeon, F.A. A crown-following approach to the automatic delineation of individual tree crowns in high spatial resolution aerial images. Can. J. Remote Sens. 1995, 21, 274–284. [Google Scholar] [CrossRef]
- Lin, C.S.; Thomson, G.; Popescu, S.C. An IPCC-compliant technique for forest carbon stock assessment using airborne LiDAR-derived tree metrics and competition Index. Remote Sens. 2016, 8, 528. [Google Scholar] [CrossRef]
- Koch, B.; Heyder, U.; Weinacker, H. Detection of individual tree crowns in airborne LiDAR data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef]
- Reitberger, J.; Schnorr, C.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LiDAR data. ISPRS J. Photogramm. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Allgöwer, B.; Nüesch, D. Clustering in airborne laser scanning raw data for segmentation of single trees. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2003, 34, 27–33. [Google Scholar]
- Vega, C.; Hamrouni, A.; El Mokhtari, S.; Morel, J.; Bock, J.; Renaud, J.P.; Bouvier, M.; Durrieu, S. PTrees: A point-based approach to forest tree extraction from LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 98–108. [Google Scholar] [CrossRef]
- Hu, X.; Chen, W.; Xu, W. Adaptive mean shift-based Identification of individual trees using airborne LiDAR Data. Remote Sens. 2017, 9, 148. [Google Scholar] [CrossRef]
- Van Leeuwen, M.; Coops, N.C.; Wulder, M.A. Canopy surface reconstruction from a LiDAR point cloud using Hough transform. Remote Sens. Lett. 2010, 1, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, J.; Yu, X.W.; Hyyppä, H.; Vastaranta, M.; Holopainen, M.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Vaaja, M.; Koskinen, J.; et al. Advances in forest inventory using airborne laser scanning. Remote Sens. 2012, 4, 1190–1207. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.J.; Hussin, Y.A. Generating pit-free canopy height models from airborne LiDAR. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Soininen, A. TerraScan User’s Guide; Terrasolid Press: Helsinki, Finland, 2000; p. 592. [Google Scholar]
- Lee, C.C.; Wang, C.K. Effect of flying altitude and pulse repetition frequency on laser scanner penetration rate for digital elevation model generation in a tropical forest. GISci. Remote Sens. 2018, 1–22. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H. Seeing the trees in the forest: Using LiDAR and multispectral data fusion with local filtering and variable window size for estimating tree height. Photogramm. Eng. Remote Sens. 2004, 70, 589–604. [Google Scholar] [CrossRef]
- Wang, Q.; Adiku, S.; Tenhunen, J.; Granier, A. On the relationship of NDVI with leaf area index in a deciduous forest site. Remote Sens. Environ. 2005, 94, 244–255. [Google Scholar] [CrossRef]
- Le Maire, G.; Marsden, C.; Nouvellon, Y.; Grinand, C.; Hakamada, R.; Stape, J.L.; Laclau, J.P. MODIS NDVI time-series allow the monitoring of Eucalyptus plantation biomass. Remote Sens. Environ. 2011, 115, 2613–2625. [Google Scholar] [CrossRef]
- Fernandez-Diaz, J.C.; Carter, W.E.; Glennie, C.; Shrestha, R.L.; Pan, Z.; Ekhtari, N.; Singhania, A.; Hauser, D.; Sartori, M. Capability assessment and performance metrics for the Titan multispectral mapping LiDAR. Remote Sens. 2016, 8, 939. [Google Scholar] [CrossRef]
- ASPRS. LAS Specification Version 1.4; American Society for Photogrammetry and Remote Sensing Press: Bethesda, MD, USA, 2011; p. 28. [Google Scholar]
- Keenan, R.J.; Reams, G.A.; Achard, F.; de Freitas, J.V.; Grainger, A.; Lindquist, E. Dynamics of global forest area: Results from the FAO global forest resources assessment 2015. For. Ecol. Manag. 2015, 352, 9–20. [Google Scholar] [CrossRef]
- Montealegre, A.L.; Lamelas, M.T.; de la Riva, J. Interpolation routines assessment in ALS-derived digital elevation models for forestry applications. Remote Sens. 2015, 7, 8631–8654. [Google Scholar] [CrossRef]
- Wulder, M.; Niemann, K.O.; Goodenough, D.G. Local maximum filtering for the extraction of tree locations and basal area from high spatial resolution imagery. Remote Sens. Environ. 2000, 73, 103–114. [Google Scholar] [CrossRef]
- Nelson, T.; Boots, B.; Wulder, M.A. Techniques for accuracy assessment of tree locations extracted from remotely sensed imagery. J. Environ. Manag. 2005, 74, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Things I have learned (so far). Am. Psychol. 1990, 45, 1304. [Google Scholar] [CrossRef]
- Kar, S.S.; Ramalingam, A. Is 30 the magic number? Issues in sample size estimation. Natl. J. Commun. Med. 2013, 4, 175–179. [Google Scholar]
- Lee, A.C.; Lucas, R.M. A LiDAR-derived canopy density model for tree stem and crown mapping in Australian forests. Remote Sens. Environ. 2007, 111, 493–518. [Google Scholar] [CrossRef]
- Palace, M.; Sullivan, F.B.; Ducey, M.; Herrick, C. Estimating tropical forest structure using a terrestrial LiDAR. PLoS ONE 2016, 11, e0154115. [Google Scholar] [CrossRef] [PubMed]
- Kahriman, A.; Gunlu, A.; Karahalil, U. Estimation of crown closure and tree density using Landsat TM satellite images in mixed forest stands. J. Indian Soc. Remote Sens. 2014, 42, 559–567. [Google Scholar] [CrossRef]
- Chrysafis, I.; Mallinis, G.; Gitas, I.; Tsakiri-Strati, M. Estimating Mediterranean forest parameters using multi seasonal Landsat 8 OLI imagery and an ensemble learning method. Remote Sens. Environ. 2017, 199, 154–166. [Google Scholar] [CrossRef]
- Mohammadi, J.; Joibary, S.S.; Yaghmaee, F.; Mahiny, A.S. Modelling forest stand volume and tree density using Landsat ETM plus data. Int. J. Remote Sens. 2010, 31, 2959–2975. [Google Scholar] [CrossRef]
- Wang, C.; Qi, J. Biophysical estimation in tropical forests using JERS-1 SAR and VNIR imagery. II. aboveground woody biomass. Int. J. Remote Sens. 2008, 29, 6827–6849. [Google Scholar] [CrossRef]



| Data | CHMFe (Csize: 1 m) | CHMFe (Csize: 0.5 m) | CHMFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 161 | 61 | 31 | 485 | 182 | 99 | 2432 | 694 | 381 | 246 | 184 | 140 | 102 | |
| 0.04 | 0.00 | 0.00 | 0.60 | 0.07 | 0.00 | 0.92 | 0.72 | 0.50 | 0.26 | 0.08 | 0.01 | 0.00 | |
| 0.21 | 0.69 | 0.83 | 0.02 | 0.14 | 0.50 | 0.00 | 0.01 | 0.03 | 0.07 | 0.14 | 0.30 | 0.48 | |
| 2.00 ★ | 4.34 | 5.36 | 10.20 | 1.68 ★ | 3.18 | 73.47 | 17.39 | 6.98 | 2.96 | 1.73 ★ | 2.30 | 3.17 | |
| Data | CHMLe (Csize: 1 m) | CHMLe (Csize: 0.5 m) | CHMLe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 232 | 104 | 49 | 775 | 314 | 169 | 3504 | 1369 | 750 | 465 | 334 | 248 | 193 | |
| 0.21 | 0.02 | 0.00 | 0.75 | 0.38 | 0.10 | 0.94 | 0.86 | 0.74 | 0.58 | 0.42 | 0.26 | 0.11 | |
| 0.07 | 0.48 | 0.75 | 0.00 | 0.02 | 0.23 | 0.00 | 0.00 | 0.00 | 0.01 | 0.02 | 0.06 | 0.13 | |
| 2.56 ★ | 3.48 | 4.90 | 18.56 | 4.53 | 2.43 ★ | 108.5 | 37.17 | 17.53 | 8.88 | 4.90 | 2.95 | 1.94 ★ | |
| Data | CHMHFe (Csize: 1 m) | CHMHFe (Csize: 0.5 m) | CHMHFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 140 | 56 | 32 | 449 | 187 | 95 | 2203 | 700 | 403 | 268 | 186 | 142 | 113 | |
| 0.04 | 0.00 | 0.00 | 0.57 | 0.10 | 0.00 | 0.91 | 0.72 | 0.52 | 0.31 | 0.12 | 0.02 | 0.01 | |
| 0.32 | 0.72 | 0.84 | 0.01 | 0.15 | 0.52 | 0.00 | 0.01 | 0.01 | 0.06 | 0.17 | 0.29 | 0.43 | |
| 2.58 ★ | 4.53 | 5.32 | 8.95 | 2.19 ★ | 3.50 | 65.36 | 16.98 | 7.53 | 3.71 | 2.51 | 2.43 ★ | 3.04 | |
| Data | CHMFe (Csize: 1 m) | CHMFe (Csize: 0.5 m) | CHMFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 0.01 | 0.11 | 0.43 | 0.07 | 0.02 | 0.09 | 0.07 | 0.01 | 0.05 | 0.09 | 0.04 | 0.14 | 0.13 | |
| 3.52 | 5.69 | 10.78 | 4.48 | 6.01 | 4.95 | 7.24 | 6.13 | 7.41 | 5.3 | 6.6 | 7.71 | 4.45 | |
| 1.48 | 1.98 | 2.66 | 1.78 | 1.24 | 1.4 | 2.08 | 1.71 | 1.5 | 1.87 | 1.62 | 1.97 | 1.66 | |
| 1.89 | 2.36 | 3.36 | 2.19 | 1.79 | 1.8 | 2.65 | 2.17 | 2.15 | 2.36 | 2.25 | 2.41 | 2.02 | |
| Data | CHMLe (Csize: 1 m) | CHMLe (Csize: 0.5 m) | CHMLe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 0.13 | 0.01 | 0.45 | 0.14 | 0.05 | 0.09 | 0.03 | 0.04 | 0.08 | 0.01 | 0.1 | 0.27 | 0.04 | |
| 6.38 | 8.07 | 14.93 | 5.7 | 5.49 | 7.39 | 5.67 | 4.79 | 5.96 | 5.6 | 6.12 | 6.58 | 4.9 | |
| 2.11 | 3.34 | 4.12 | 1.85 | 2.14 | 2.2 | 2.05 | 1.9 | 1.86 | 2.07 | 1.98 | 2.36 | 1.85 | |
| 2.6 | 3.88 | 4.91 | 2.34 | 2.67 | 2.85 | 2.59 | 2.26 | 2.3 | 2.44 | 2.47 | 2.91 | 2.28 | |
| Data | CHMHFe (Csize: 1 m) | CHMHFe (Csize: 0.5 m) | CHMHFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| 0.14 | 0.01 | 0.32 | 0.05 | 0.02 | 0.14 | 0.08 | 0.15 | 0.15 | 0.03 | 0.01 | 0.01 | 0.01 | |
| 12.65 | 6 | 8.81 | 6.9 | 5.84 | 6.75 | 5.29 | 4.84 | 5 | 7.23 | 6.65 | 6.44 | 5.09 | |
| 2.03 | 2.39 | 3.73 | 1.71 | 1.85 | 2.09 | 1.94 | 1.86 | 1.58 | 1.84 | 1.85 | 1.82 | 1.83 | |
| 3.08 | 2.8 | 4.94 | 2.19 | 2.43 | 2.65 | 2.34 | 2.26 | 1.92 | 2.43 | 2.52 | 2.39 | 2.28 | |
| Data | CHMFe (Csize: 1 m) | CHMFe (Csize: 0.5 m) | CHMFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| −0.07 | 0.01 | −0.02 | −0.22 | −0.08 | −0.01 | −0.75 | −0.19 | −0.11 | −0.06 | −0.04 | −0.04 | 0.00 | |
| 1.58 | 0.32 | 0.32 | 4.59 | 1.74 | 0.71 | 18.48 | 5.56 | 3.08 | 1.87 | 1.40 | 1.12 | 0.54 | |
| −1.55 | −0.29 | −0.24 | −3.28 | −1.58 | −0.84 | −5.20 | −3.99 | −2.28 | −1.33 | −0.91 | −0.82 | −0.30 | |
| 0.18 | 0.16 | 0.21 | 0.20 | 0.15 | 0.12 | 0.24 | 0.19 | 0.17 | 0.18 | 0.17 | 0.23 | 0.14 | |
| 0.15 | 0.17 | 0.21 | 0.15 | 0.10 | 0.11 | 0.14 | 0.12 | 0.11 | 0.12 | 0.11 | 0.14 | 0.14 | |
| 2.40 | 2.18 | 2.83 | 2.60 | 2.17 | 1.74 | 2.91 | 2.64 | 2.60 | 2.56 | 2.60 | 3.14 | 1.90 | |
| Data | CHMLe (Csize: 1 m) | CHMLe (Csize: 0.5 m) | CHMLe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| −0.09 | −0.04 | 0.00 | −0.33 | −0.15 | −0.07 | −1.74 | −0.63 | −0.27 | −0.21 | −0.16 | −0.16 | −0.11 | |
| 1.78 | 0.74 | 0.15 | 6.29 | 2.55 | 1.18 | 31.47 | 11.32 | 5.55 | 3.88 | 2.95 | 2.53 | 1.89 | |
| 0.33 | 0.23 | 0.55 | −0.20 | 0.59 | 0.83 | −8.88 | 0.01 | 0.60 | −0.20 | −0.73 | −0.97 | −0.78 | |
| 0.19 | 0.29 | 0.35 | 0.17 | 0.17 | 0.21 | 0.21 | 0.19 | 0.16 | 0.16 | 0.17 | 0.17 | 0.14 | |
| 0.16 | 0.18 | 0.17 | 0.15 | 0.15 | 0.15 | 0.16 | 0.12 | 0.15 | 0.16 | 0.12 | 0.12 | 0.10 | |
| 2.53 | 3.56 | 4.88 | 2.42 | 2.35 | 2.82 | 2.76 | 2.37 | 2.17 | 2.19 | 2.12 | 2.02 | 1.76 | |
| Data | CHMHFe (Csize: 1 m) | CHMHFe (Csize: 0.5 m) | CHMHFe (Csize: 0.2 m) | ||||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | |
| −0.04 | 0.01 | 0.00 | −0.12 | −0.09 | −0.03 | −0.82 | −0.18 | −0.14 | −0.13 | −0.07 | −0.05 | −0.04 | |
| 1.04 | 0.16 | 0.10 | 3.49 | 1.79 | 0.78 | 18.26 | 5.05 | 3.46 | 2.58 | 1.74 | 1.28 | 1.03 | |
| −0.38 | 0.14 | 0.19 | −2.03 | −1.21 | −0.53 | −8.28 | −1.56 | −2.57 | −1.92 | −1.54 | −0.91 | −0.99 | |
| 0.23 | 0.20 | 0.28 | 0.19 | 0.18 | 0.25 | 0.22 | 0.20 | 0.17 | 0.18 | 0.21 | 0.21 | 0.22 | |
| 0.16 | 0.11 | 0.17 | 0.13 | 0.18 | 0.16 | 0.14 | 0.14 | 0.13 | 0.15 | 0.17 | 0.16 | 0.17 | |
| 3.47 | 2.24 | 3.71 | 2.71 | 2.59 | 3.45 | 2.69 | 2.55 | 2.39 | 2.59 | 3.04 | 2.92 | 3.00 | |
| Study | Remote Sensing Data | Forest Type | |
|---|---|---|---|
| This study | airborne LiDAR (Optech ALTM 3070) | tropical forest | 1.68 * |
| Lee and Lucas [40] | airborne LiDAR (Optech ALTM 1020) | white cypress pine | 1.33 * |
| Palace, et al. [41] | terrestrial LiDAR (FARO Focus 3D) | tropical forest | 1.53 * |
| Kahriman, et al. [42] | optical satellite (Landsat TM) | pine and beech | 0.83 * |
| Chrysafis, et al. [43] | optical satellite (Landsat 8 OLI) | black pine and oaks | 2.57 * |
| Mohammadi, et al. [44] | optical satellite (Landsat ETM+) | hornbeam and oak | 1.70 * |
| Wang and Qi [45] | SAR satellite (JERS-1) | deciduous forests | 1.61 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.-C.; Wang, C.-K. Estimating Stand Density in a Tropical Broadleaf Forest Using Airborne LiDAR Data. Forests 2018, 9, 475. https://doi.org/10.3390/f9080475
Lee C-C, Wang C-K. Estimating Stand Density in a Tropical Broadleaf Forest Using Airborne LiDAR Data. Forests. 2018; 9(8):475. https://doi.org/10.3390/f9080475
Chicago/Turabian StyleLee, Chung-Cheng, and Chi-Kuei Wang. 2018. "Estimating Stand Density in a Tropical Broadleaf Forest Using Airborne LiDAR Data" Forests 9, no. 8: 475. https://doi.org/10.3390/f9080475





