Tamarix microRNA Profiling Reveals New Insight into Salt Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Design
2.2. Peroxidase (POD) Enzyme Activity
2.3. Small RNA Deep Sequencing
2.4. Conserved and Novel MicroRNAs Identification
2.5. Differentially Expressed MicroRNA Identification
2.6. Target Genes Prediction
2.7. MicroRNA Expression Validation by qRT-PCR
3. Results
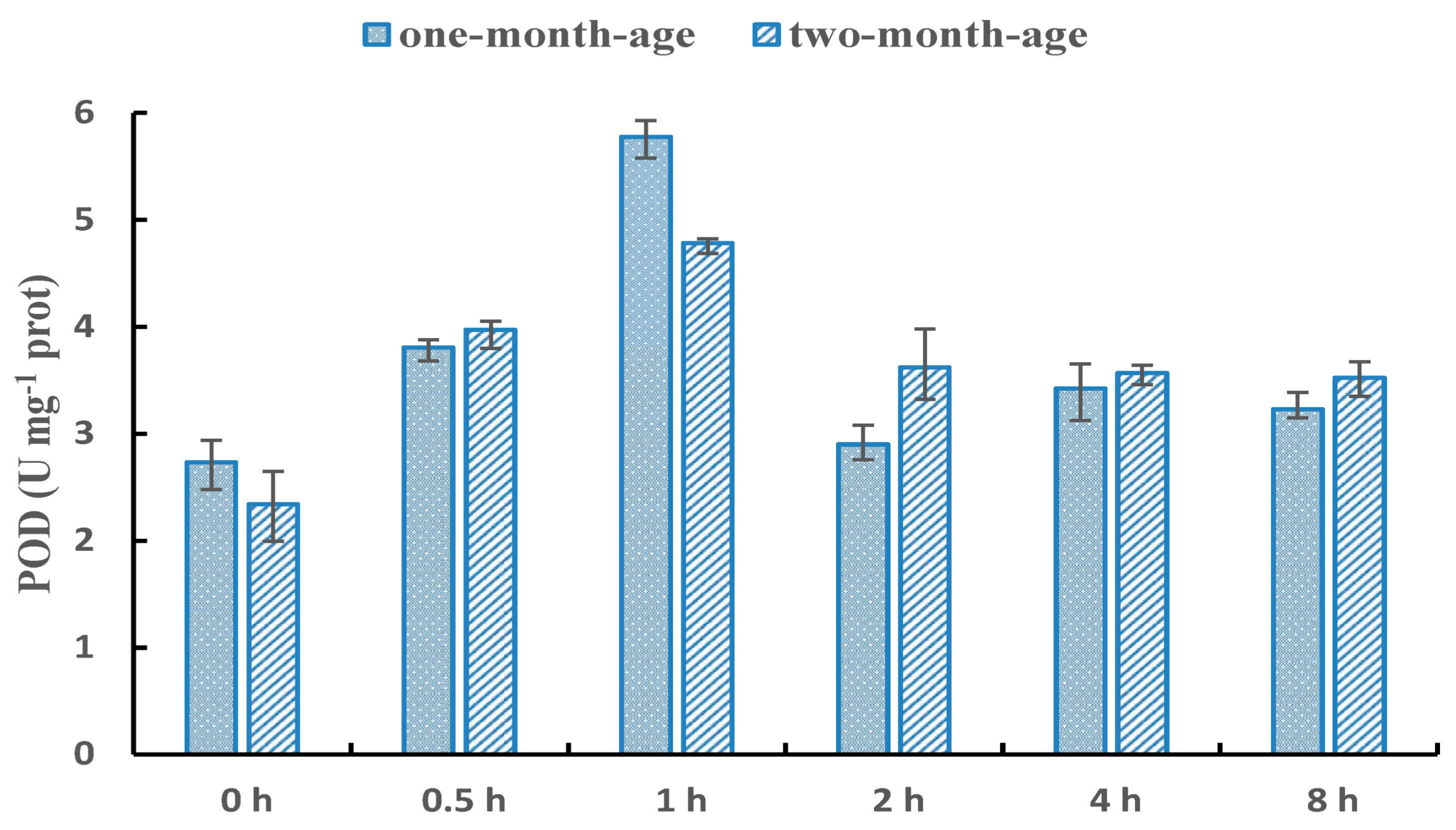
3.1. Selection of NaCl Treatment Method
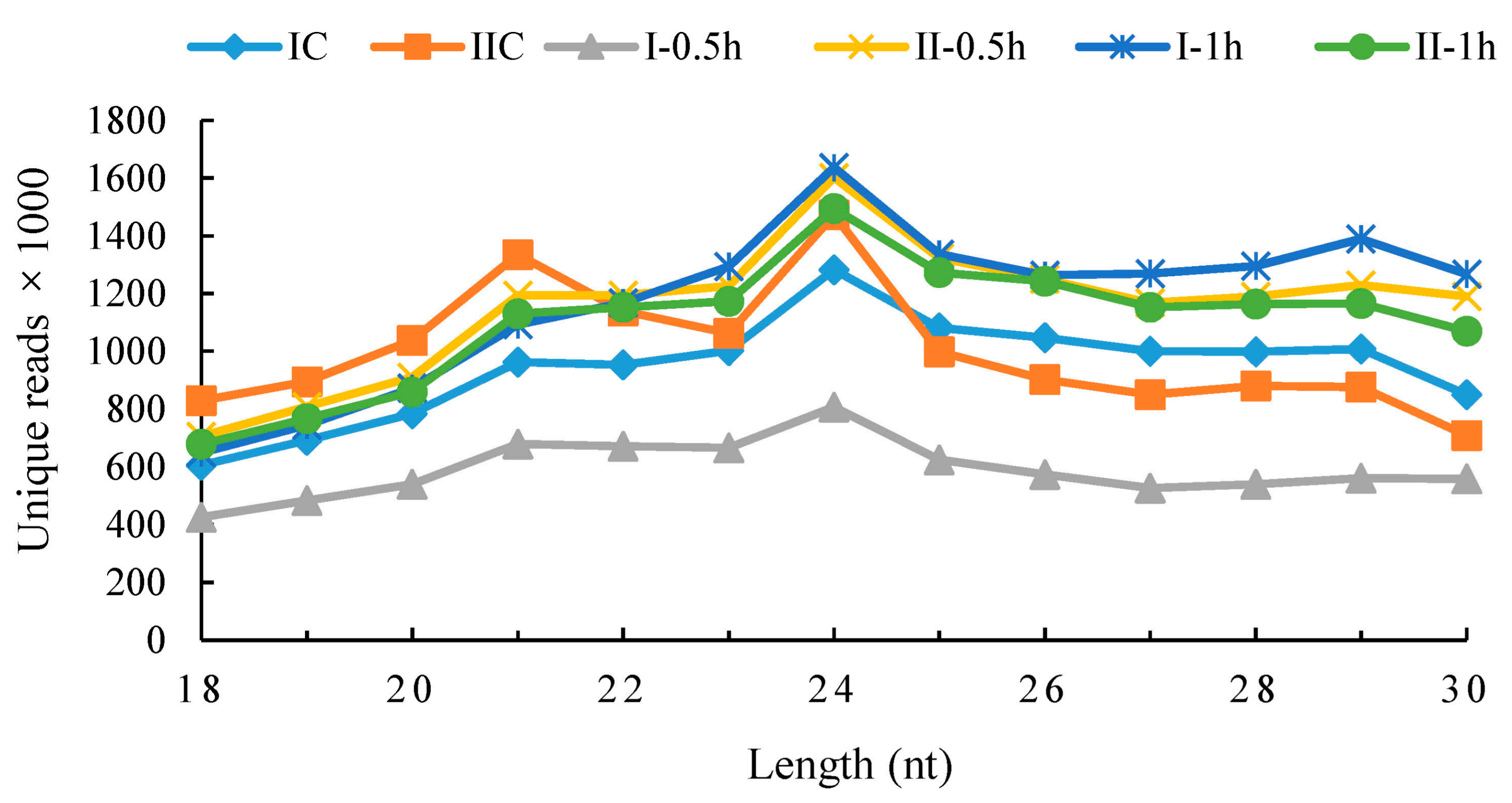
3.2. Small RNAs Generated from Deep Sequencing
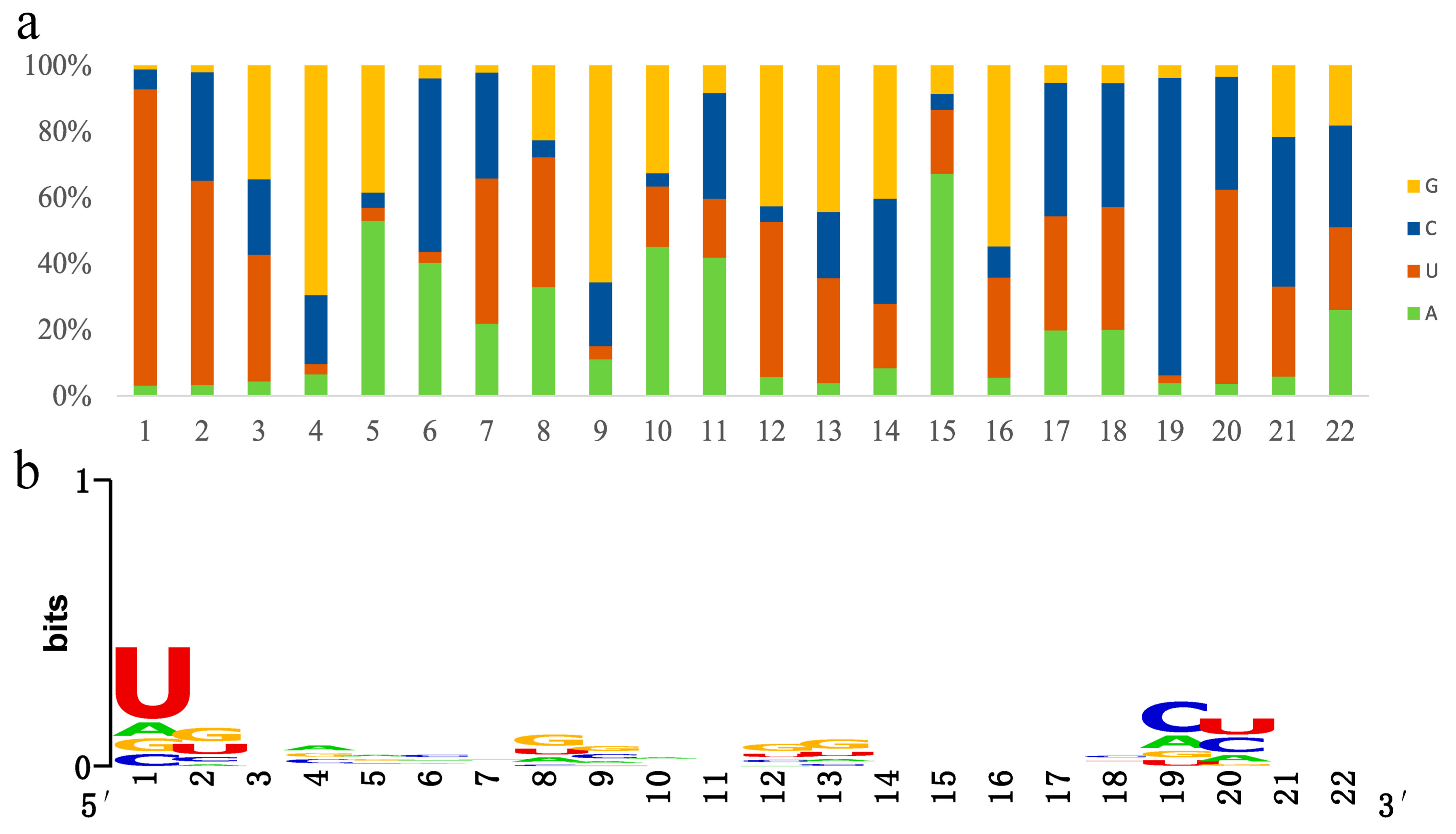
3.3. Conserved and Novel MicroRNA Identification
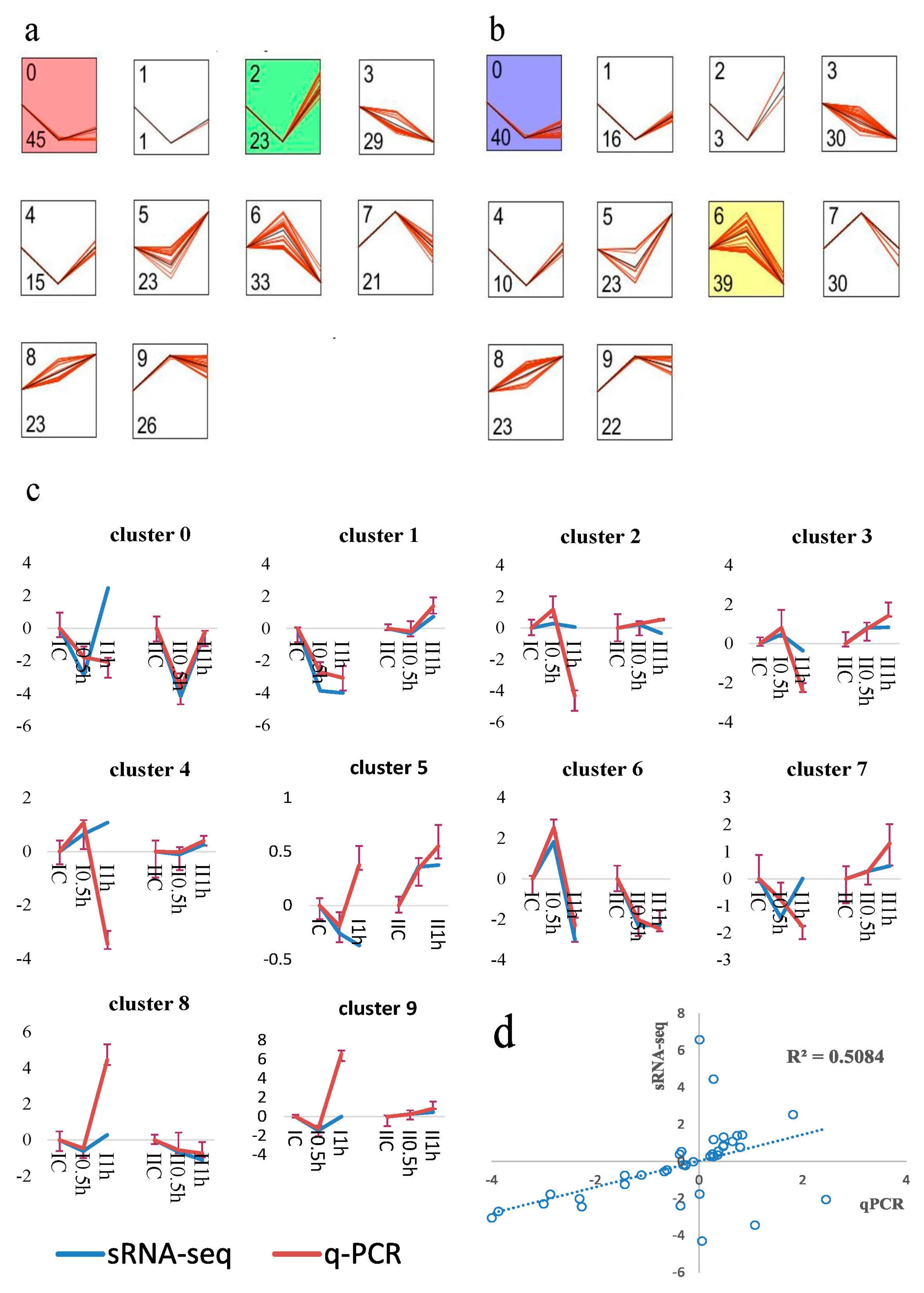
3.4. MicroRNA Profile Building and Time-Course Analysis
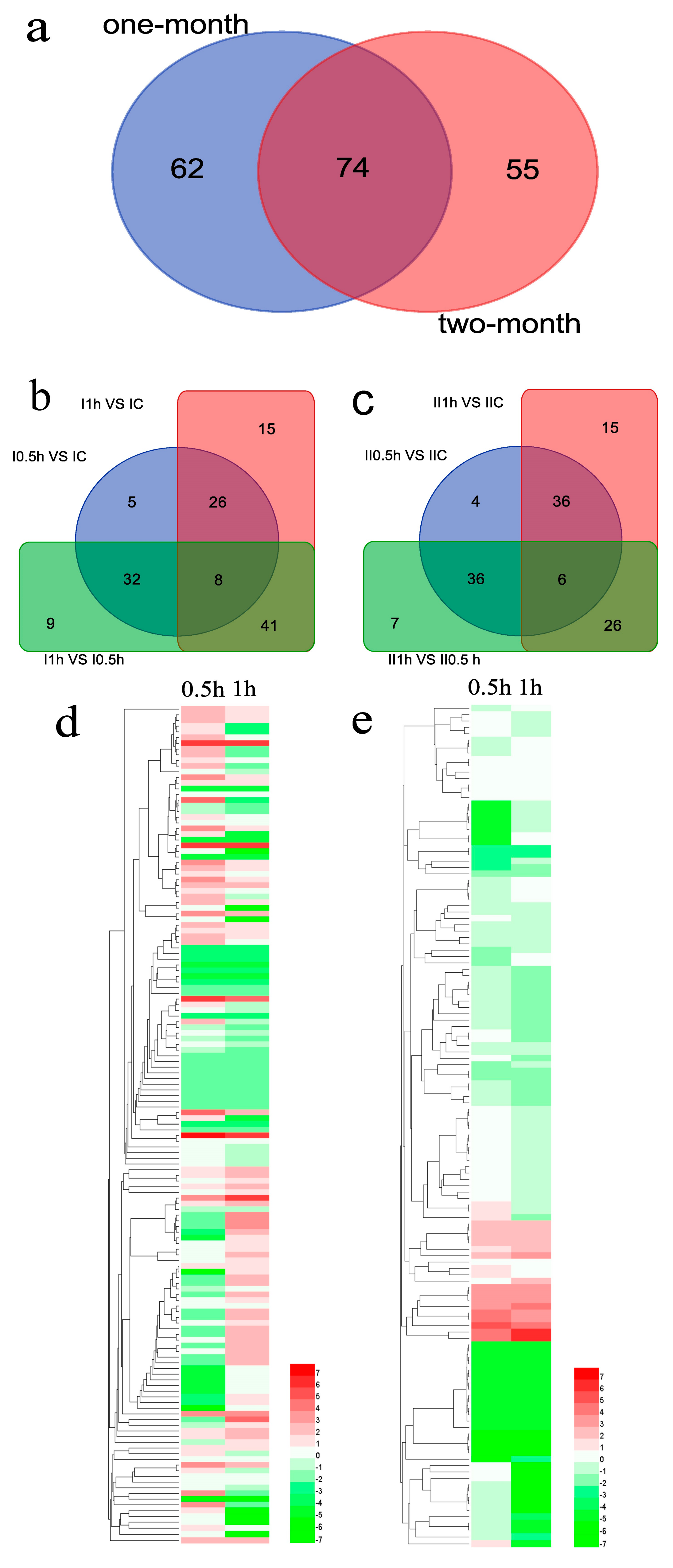
3.5. Differentially Expressed MicroRNAs Identification
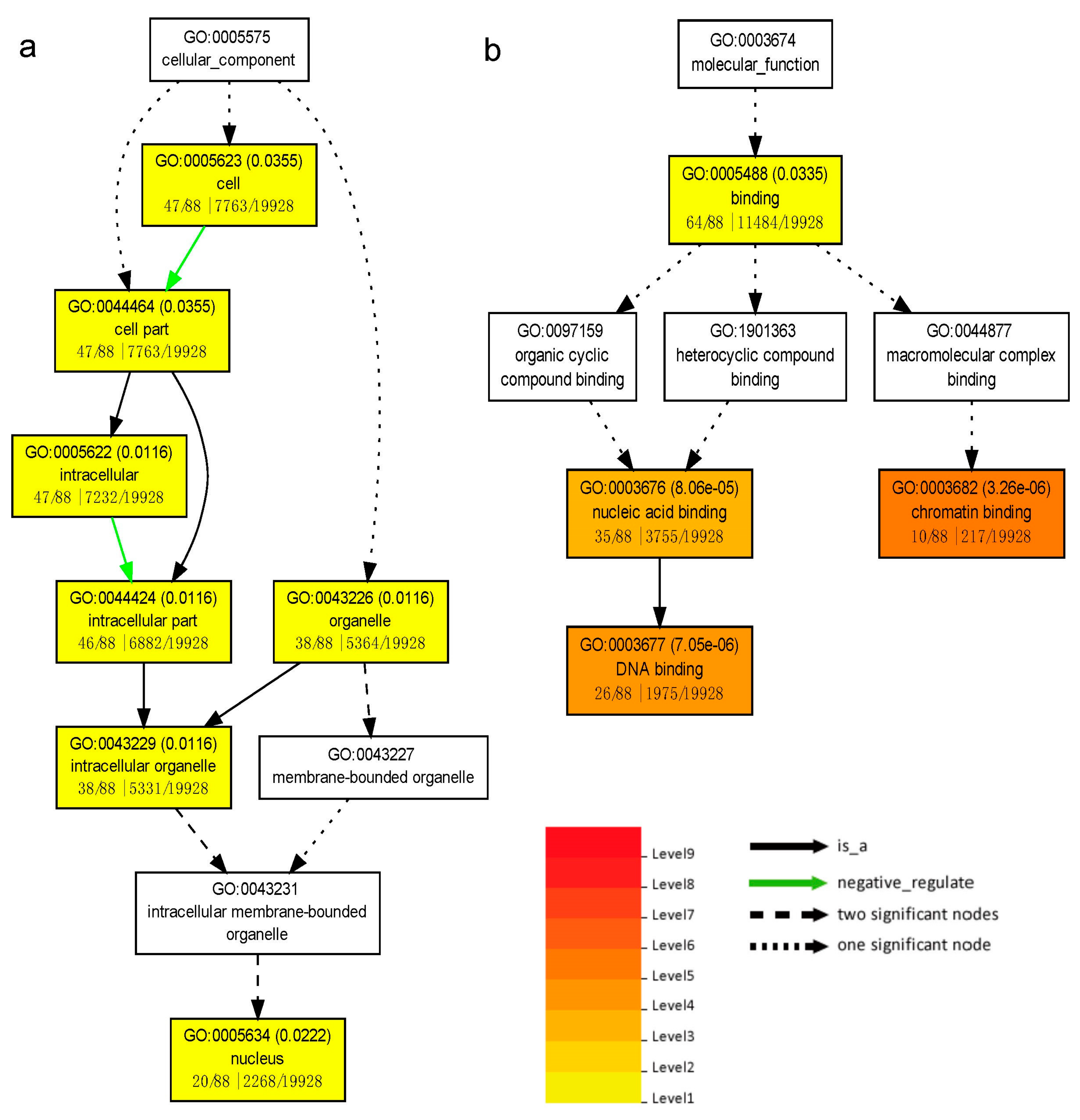
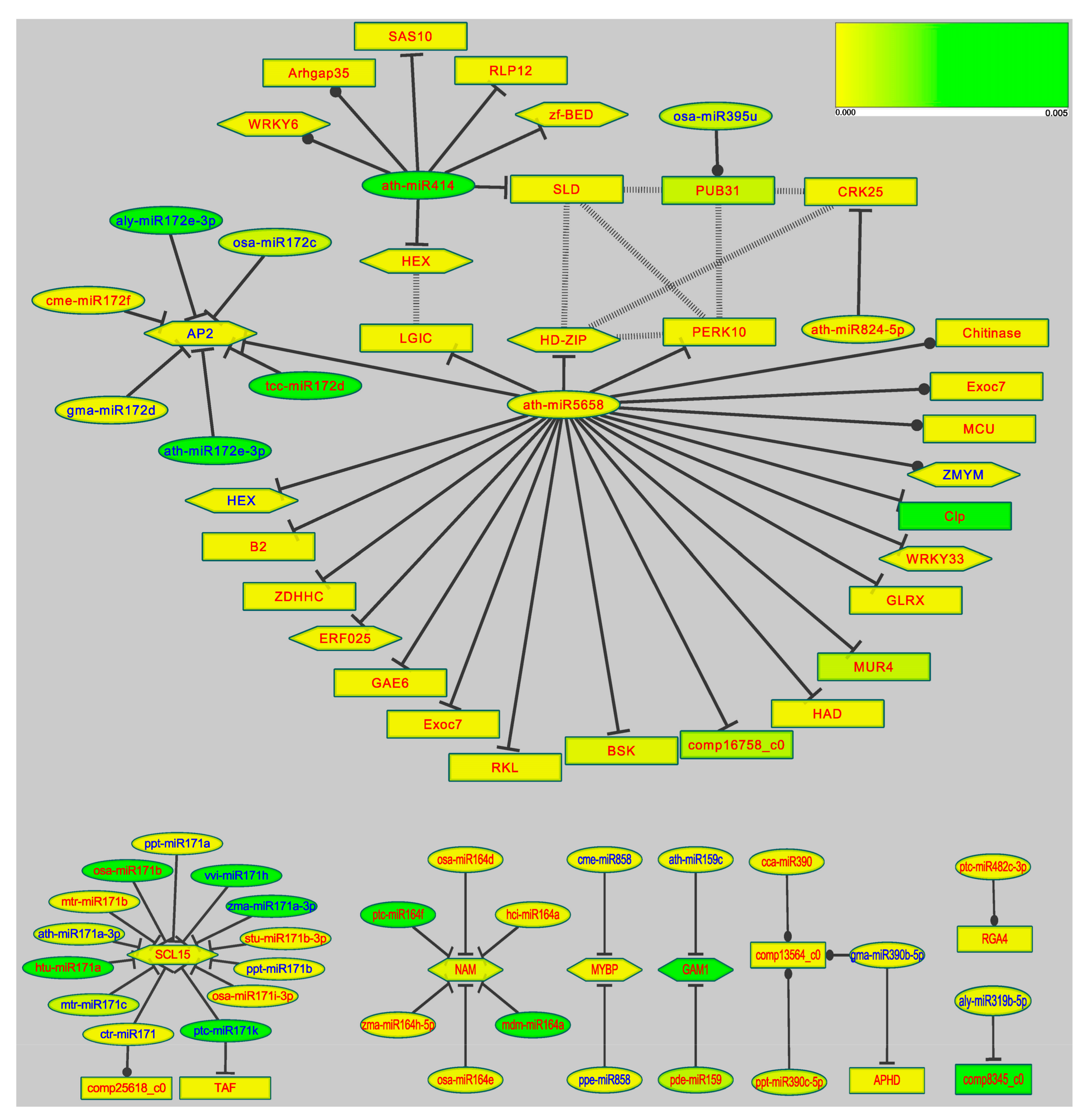
3.6. Target Genes Prediction and MicroRNA Regulatory Network
4. Discussion
4.1. Strategy for Time-Course Experimental Design
4.2. The miRNA Regulation Network of Tamarix under Stress
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Brousse, C.; Liu, Q.; Beauclair, L.; Deremetz, A.; Axtell, M.J.; Bouche, N. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014, 42, 5270–5279. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Reichel, M.; Li, Y.; Millar, A.A. The functional scope of plant microRNA-mediated silencing. Trends Plant Sci. 2014, 19, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. BBA-Gene Regul. Mech. 2008, 1779, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot.-Lond. 2009, 103, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Xie, F., Jr.; Stewart, C.N.; Taki, F.A.; He, Q.; Liu, H.; Zhang, B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol. J. 2014, 12, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant salt stress: Adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S. Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front. Plant Sci. 2017, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Soda, N.; Sharan, A.; Gupta, B.K.; Singlapareek, S.L.; Pareek, A. Evidence for nuclear interaction of a cytoskeleton protein (OsIFL) with metallothionein and its role in salinity stress tolerance. Sci. Rep.-UK 2016, 6, 34762. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Jin, Z.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep.-UK 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Dugas, D.V.; Bartel, B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant. Mol. Biol. 2008, 67, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Kawa, D.; Testerink, C. Regulation of mRNA decay in plant responses to salt and osmotic stress. Cell. Mol. Life Sci. 2017, 74, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Khare, T.; Shriram, V.; Wani, S.H. Plant small RNAs: The essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Rep. 2018, 37, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.; Liu, G.; Yang, C.; Jiang, J.; Li, H. Expression profiling of salinity-alkali stress responses by large-scale expressed sequence tag analysis in Tamarix hispid. Plant Mol. Biol. 2008, 66, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Liu, G.; Liu, Y.; Zheng, L.; Nie, X.; Wang, Y. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 2013, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, C.; Wang, Y.; Guo, Y.; Zhao, Y.; Yang, C.; Gao, C. Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of Cadmium stress. Sci. Rep.-UK 2016, 6, 18752. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.P.; Wang, Y.C.; Liu, G.F.; Jiang, J.; Zhang, G.D. Study on expression of genes in Tamarix androssowii under NaHCO3 stress using gene chip technology. Sheng Wu Gong Cheng Xue Bao 2005, 21, 220–226. [Google Scholar] [PubMed]
- Wang, C.; Gao, C.; Wang, L.; Zheng, L.; Yang, C.; Wang, Y. Comprehensive transcriptional profiling of NaHCO3-stressed Tamarix hispida roots reveals networks of responsive genes. Plant Mol. Biol. 2014, 84, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, H. Designing microarray and RNA-Seq experiments for greater systems biology discovery in modern plant genomics. Mol. Plant 2015, 8, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; Sunkar, R.; Zheng, Y.; Ji, B.; Al-Yahyai, R.; Farooq, S.A. A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front. Plant Sci. 2015, 6, 946. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zang, B.; Yao, H.S.; Huang, M.R. Isolation of high quality RNA and molecular manipulations with various tissues of Populus. Russ. J. Plant Physl. 2009, 56, 716–719. [Google Scholar] [CrossRef]
- Wang, J.; Xu, M.; Gu, Y.; Xu, L. Differentially expressed gene analysis of Tamarix chinensis provides insights into NaCl-stress response. Trees-Struct. Funct. 2017, 31, 645–658. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11–17. [Google Scholar]
- Griffiths-Jones, S. miRBase: The microRNA Sequence Database; Humana Press: New York, NY, USA, 2006; Volume 342, pp. 129–138. [Google Scholar]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Moxon, S.; Schwach, F.; Dalmay, T.; MacLean, D.; Studholme, D.J.; Moulton, V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 2008, 24, 2252–2253. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 99, 138–148. [Google Scholar]
- Wu, H.J.; Ma, Y.K.; Chen, T.; Wang, M.; Wang, X.J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Guo, C. Effects of salt stress on SOD and POD activities in three bamboos. J. Jiangsu Forestryence Technol. 1995, 22, 11–14. [Google Scholar]
- Wan, L.; Wang, F.; Guo, X.; Lu, S.; Qiu, Z.; Zhao, Y.; Zhang, H.; Lin, J. Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol. 2012, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Q.; Zhou, H.; Ni, F.; Wu, X.; Qi, Y. Rice microRNA effector complexes and targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, W.; Zhang, P.; Zhang, J.; Liu, L. Study on the salt resistance of conifer Tamarisk in hydroponic experiment. J. Tianjin Univ. Technol. 2016, 32, 42–45. [Google Scholar]
- Liu, K.; Deng, C.; Hao, J. Physiological response of Tamarix austromongolica to NaCl stress. Guangdong Agric. Sci. 2012, 10, 38–42. [Google Scholar]
- Hansen, K.D.; Wu, Z.; Irizarry, R.A.; Leek, J.T. Sequencing technology does not eliminate biological variability. Nat. Biotechnol. 2011, 29, 572–573. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; White, K.P. RNA-seq differential expression studies: More sequence or more replication? Bioinformatics 2014, 30, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Schurch, N.J.; Schofield, P.; Gierlinski, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gierliński, M.; Cole, C.; Schofield, P.; Schurch, N.J.; Sherstnev, A. Statistical models for RNA-seq data derived from a two-condition 48-replicate experiment. Bioinformatics 2015, 31, 3625–3630. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, L.M.; Lopiano, K.K.; Morse, A.M.; Amin, V.; Oberg, A.L.; Young, L.J.; Nuzhdin, S.V. RNA-seq: Technical variability and sampling. BMC Genom. 2011, 12, 293. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Zhang, S.; Korpelainen, H.; Li, C. Physiological and transcriptional responses of two contrasting Populus clones to nitrogen stress. Tree Physiol. 2016, 36, 628–642. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Hwang, E.; Shin, S.; Yu, B.; Byun, M.; Kwon, H. miR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 2011, 54, 43–48. [Google Scholar] [CrossRef]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef] [PubMed]
- Breakfield, N.W.; Corcoran, D.L.; Petricka, J.J.; Shen, J.; Sae-Seaw, J.; Rubio-Somoza, I.; Weigel, D.; Ohler, U.; Benfey, P.N. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 2012, 22, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Zhao, H.; Zhao, Y.; Cheng, B.; Xiang, Y. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014, 9, e87156. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Vargas, S.; Sanchez-Garcia, A.; Martinez-Rivas, J.M.; Prieto, J.A.; Randez-Gil, F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl. Environ. Microb. 2006, 73, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef] [PubMed]
| Samples ID | Tissue | Age | Treatment |
|---|---|---|---|
| IC | primary roots from cuttings of different age | Group I: One-month-old cuttings | water for 1 h, control group |
| I-0.5h | 2% salt for 0.5 h | ||
| I-1h | 2% salt for 1 h | ||
| IIC | Group II: Two-month-old cuttings | Water for 1 h, control group | |
| II-0.5h | 2% salt for 0.5 h | ||
| II-1h | 2% salt for 1 h |
| Small RNA Library | Roots of 2% NaCl Treated Cuttings | Type of Analysis | |||||
|---|---|---|---|---|---|---|---|
| Group I | Group II | ||||||
| IC | I-0.5h | I-1h | IIC | II-0.5h | II-1h | ||
| Clean reads | 20,304,895 (1.038 G) | 12,017,139 (0.626 G) | 27,490,082 (1.426 G) | 21,379,294 (1.093 G) | 24,151,563 (1.231 G) | 23,758,579 (1.215 G) | Deep sequencing |
| Clean reads of small RNA | 12,268,690 | 7,656,408 | 15,286,485 | 12,991,605 | 14,999,466 | 14,327,078 | Length selection |
| Mapped small RNA reads | 8,450,784 (68.88%) | 3,535,105 (46.17%) | 6,984,424 (45.69%) | 7,002,413 (53.9%) | 8,045,446 (53.64%) | 7,872,758 (54.95%) | Map to reference |
| rRNA and tRNA | 1,531,042 (18.12%) | 631,272 (17.86%) | 1,352,577 (19.37%) | 1,242,869 (17.75%) | 1,378,558 (17.13%) | 1,294,286 (16.44%) | BLAST |
| snRNA and snoRNA | 30,146 (0.36%) | 9131 (0.26%) | 23,888 (0.34%) | 23,057 (0.33%) | 37,014 (0.46%) | 22,183 (0.28%) | |
| Conserved miRNA reads | 41,979 (0.50%) | 29,239 (0.83%) | 31,756 (0.45%) | 100,486 (1.44%) | 57,517 (0.71%) | 45,494 (0.58%) | BLAST |
| Conserved unique miRNA | 180 | 158 | 167 | 207 | 187 | 170 | |
| Novel miRNA reads | 609 (0.01%) | 491 (0.01%) | 522 (0.01%) | 1282 (0.02%) | 848 (0.01%) | 642 (0.01%) | Prediction |
| Novel miRNA | 12 | 12 | 14 | 15 | 13 | 14 | |
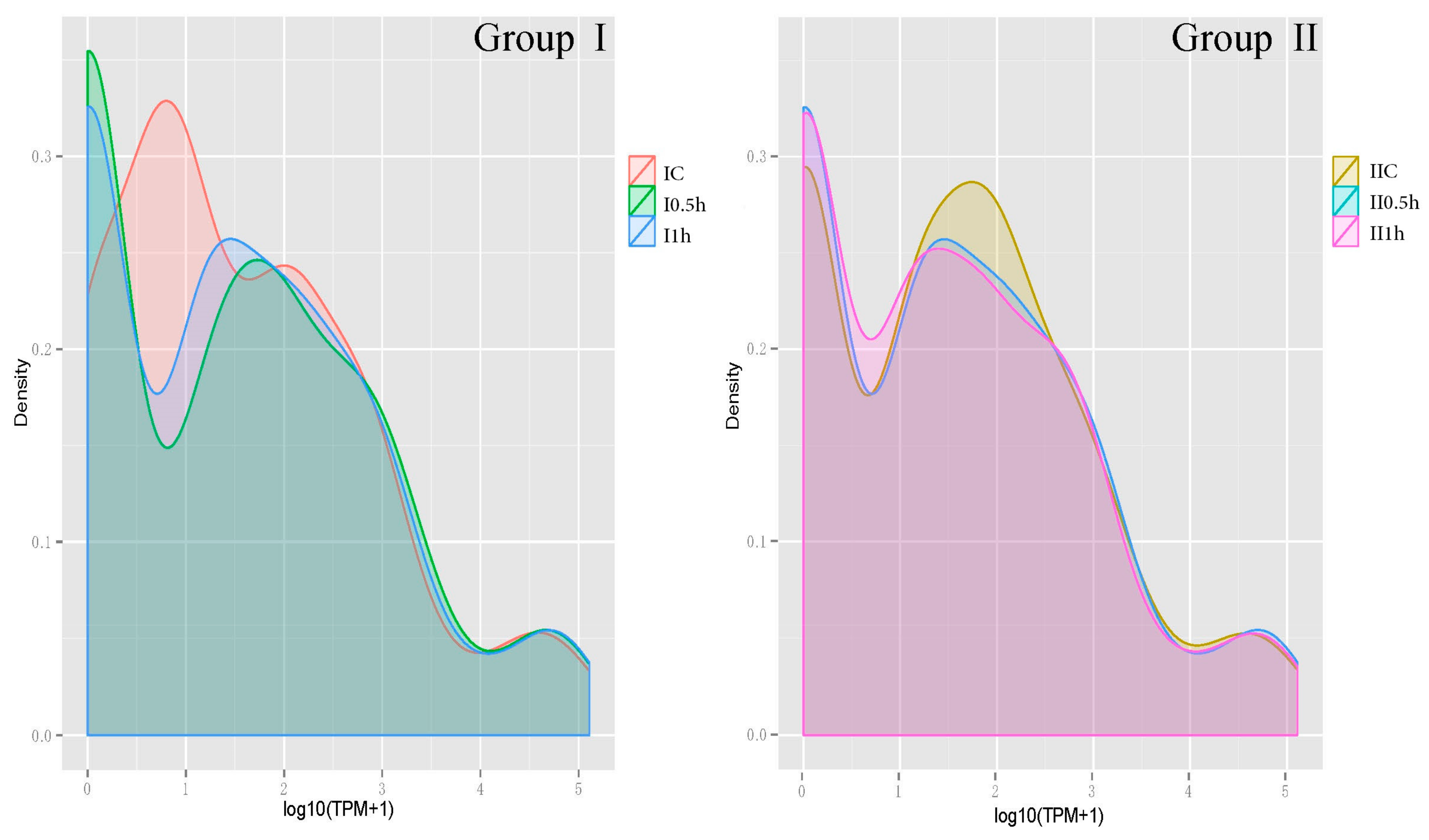
| TPM Interval | IC | IIC | I-0.5h | II-0.5h | I-1h | II-1h | Mean |
|---|---|---|---|---|---|---|---|
| 0–15 | 128 (47.58%) | 109 (40.52%) | 125 (46.47%) | 125 (46.47%) | 128 (47.58%) | 118 (42.87%) | 122 (45.41%) |
| 15–60 | 28 (10.41%) | 43 (15.99%) | 30 (11.15%) | 30 (11.15%) | 35 (13.01%) | 43 (15.99%) | 35 (12.95%) |
| >60 | 113 (42.01%) | 117 (43.49%) | 114 (42.38%) | 114 (42.38%) | 106 (39.41%) | 108 (40.15%) | 112 (41.64) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Xu, M.; Li, Z.; Ye, Y.; Rong, H.; Xu, L.-a. Tamarix microRNA Profiling Reveals New Insight into Salt Tolerance. Forests 2018, 9, 180. https://doi.org/10.3390/f9040180
Wang J, Xu M, Li Z, Ye Y, Rong H, Xu L-a. Tamarix microRNA Profiling Reveals New Insight into Salt Tolerance. Forests. 2018; 9(4):180. https://doi.org/10.3390/f9040180
Chicago/Turabian StyleWang, Jianwen, Meng Xu, Zhiting Li, Youju Ye, Hao Rong, and Li-an Xu. 2018. "Tamarix microRNA Profiling Reveals New Insight into Salt Tolerance" Forests 9, no. 4: 180. https://doi.org/10.3390/f9040180





