1. Introduction
Tree biomass is the foundation of forest ecosystems, and has been subject to research for over a century. In recent years, estimating tree total and component biomass has been greatly increased (e.g., [
1,
2,
3,
4]) because they are needed when predicting net primary production (NPP) for different stands or regions, and estimating total carbon and fluxes in forest ecosystems [
5,
6,
7,
8].
Total tree biomass is commonly divided into different components according to their physiological functions such as stems, roots, branches, and foliage. Although directly measuring the actual weight of each component is undoubtedly the most accurate method, it is destructive, time consuming, and costly. Thus, using allometric equations to develop biomass models is considered a better and more feasible approach to estimating tree-level biomass [
9,
10,
11]. Over the last few decades, more than 2600 biomass models have been developed for more than 100 species around the world [
12,
13,
14,
15,
16,
17], especially for the aboveground biomass. Belowground biomass is an important component of forest biomass that demands more investigation and study. However, this component is not tracked in many forest inventories, because extracting tree roots is expensive and time consuming, and the techniques for measuring root biomass are poorly developed compared to stem, branch and foliage biomass [
11,
18]. Until now, only few models have been made available for tree belowground or root biomass. For these biomass models published in the literature, tree diameter at breast height (
D) is the most commonly used and reliable predictor for total, sub-total, and components biomass [
11,
19,
20,
21,
22,
23]. Although other tree variables have been investigated as potential predictors for tree biomass models, tree total height is considered the best, in addition to tree diameter, to significantly improve model fitting and performance [
24,
25,
26,
27,
28].
The allometric equation
Y =
a ×
Xb is a mathematical function commonly used for tree biomass modeling. In practice, logarithmic transformation is routinely used to fit the allometric equation using ordinary least squares regression [
14,
29,
30]. However, some researchers suggest that data analysis and modeling should be performed on the original data of measurement via nonlinear regression [
10,
27]. The choice between linear regression on log-transformed data (LR) or nonlinear regression on original data (NLR) depends on the model error structure. NLR fits the original data by nonlinear least squares assuming an additive error structure for the allometric equation, whereas LR fits the log-transformed data assuming the underlying power function with a multiplicative error structure. To facilitate the objective determination on the model error structures, Bi et al. [
10] compared the predictive performance of the two model specifications for a system of additive biomass equations using the ratio of their mean squared errors. Xiao et al. [
31] and Ballantyne [
32] outlined the approach of likelihood analysis to evaluating and comparing model error structures, which was recently used for tree biomass [
30,
33]. Compared with the MSE ratio approach, the likelihood analysis is considered to be consistent with core statistical principles, and more suitable in determining model error structures [
32].
To date, biomass equations for estimating tree total, sub-total, and component biomass can be classified into non-additive and additive in nature. Non-additive biomass equations fit the biomass data of total, sub-total and components separately. Consequently, the sum of model predictions from component biomass models may not be equal to the model prediction from the total biomass model. In contrast, additive biomass equations fit the biomass data of total, sub-total and components simultaneously to account for the inherent correlations among biomass components measured on the same sample trees. Thus, the sum of biomass predictions from the component biomass equations is equal to the biomass prediction from the total or sub-total equation [
34,
35]. To achieve the additivity for a system of biomass equations, different model parameter estimation methods have been suggested for both linear and nonlinear biomass models [
10,
34,
35,
36]. In particular, seemingly unrelated regression (SUR) and nonlinear seemingly unrelated regression (NSUR) are more general and flexible, and have become more popular as the parameter estimation method for linear and nonlinear biomass equations [
37,
38,
39,
40,
41].
Past research has indicated that the origin of forest stands may influence the biomass estimation [
26,
42,
43,
44]. Generally, if a tree species exists in both plantation and natural forests, the biomass models should be developed separately for each forest origin. In the forest regions of northeastern China, Korean pine (
Pinus koraiensis Sieb. et Zucc.), larch (
Larix gmelinii (Rupr.) Kuzen.) and Mongoli
an pine (
Pinus sylvestris var.
mongolica Litv.) are three major species growing in plantation forests. To date, Wang [
11] and Dong et al. [
33] developed biomass equations for Korean pine and larch in northeastern China. However, Wang’s [
11] biomass data were collected from a limited forest region with a relatively small sample size for each species. The biomass data of Dong et al. [
33] came from natural forest stands. For Mongolian pine, there was very limited information available on aboveground and belowground or root biomass. Thus, the objectives of this study were: (1) to analyze the biomass partitioning for the three species; (2) to examine which model error structure is more suitable for the allometric biomass relationships; (3) to develop two additive systems of biomass equations for the three species; and (4) to validate the performance of the biomass models through the jackknifing technique across the classes of tree diameter.
3. Results
3.1. Biomass Partitioning
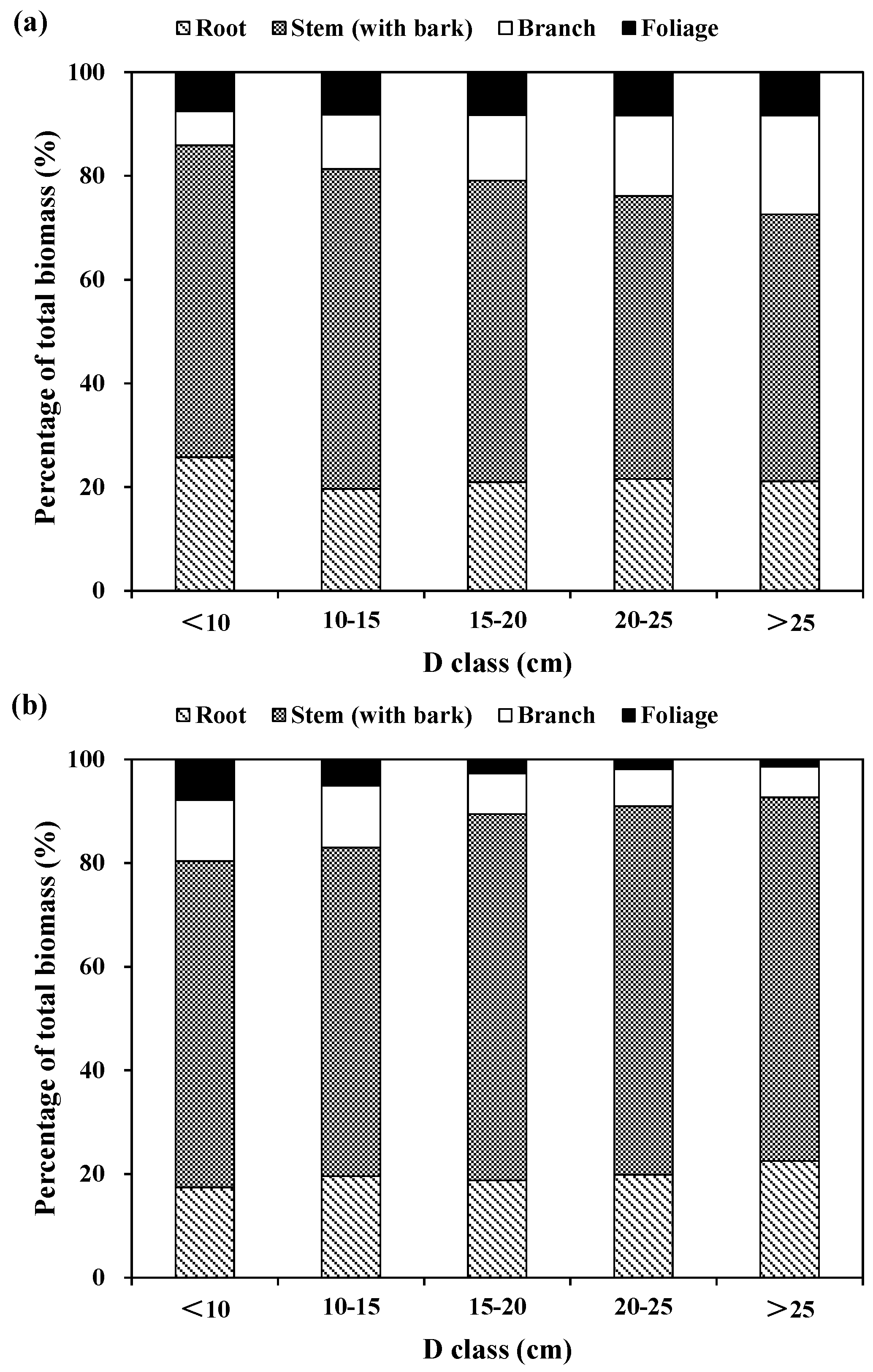
The relative partitioning of tree components to total tree biomass was also computed for the three species across tree diameter classes (
Figure 2). For Korean pine, the relative contribution of stem (with bark) biomass to total biomass increased from about 50% for large diameter classes to 60% for small and medium diameter classes. The proportion of root biomass is about 25% for the small diameter class and 20% for the medium and large diameter class. The proportion of branch biomass increased from 7% for the small diameter class and to 19% for the large diameter class. However, the relative contribution of foliage biomass increased marginally with tree diameter classes, because the differences of this relative contribution were not significant among tree diameter classes (
Figure 2a). For larch, the proportions of stem and root biomass increased from 63% and 17% for the small diameter class, and to 70% and 21% for medium and large diameter classes, respectively. However, the proportions of branch and foliage biomass decreased from about 12% and 7% for the small diameter class to 6% and 2% for the large diameter class, respectively (
Figure 2b). For Mongolian pine, the proportions of root and foliage biomass decreased from 25% and 9% for the small diameter class, and to 15% and 6% for the large diameter class, respectively; however, the proportions of stem biomass increased from 52% for the small diameter class and to 68% for large diameter class. The proportion of branch biomass was greater for the smallest and largest diameter classes, and lesser for the medium diameter class (
Figure 2c).
In general, stem biomass had the largest relative contribution to total biomass, while foliage biomass had the smallest relative proportion for the three plantation species. For Korean pine, the proportion was 57% for stem, 22% for root, 13% for branch, and 8% for foliage. For larch, the proportion was 67% for stem, 20% for root, 9% for branch, and 4% for foliage. For Mongolian pine, the proportion was 63% for stem, 20% for root, 10% for branch, and 7% for foliage. On average, the aboveground (i.e., the sum of stems, branches and foliage) biomass was usually above 75% of the total biomass, while the belowground biomass (i.e., roots) was below 25% of the total biomass.
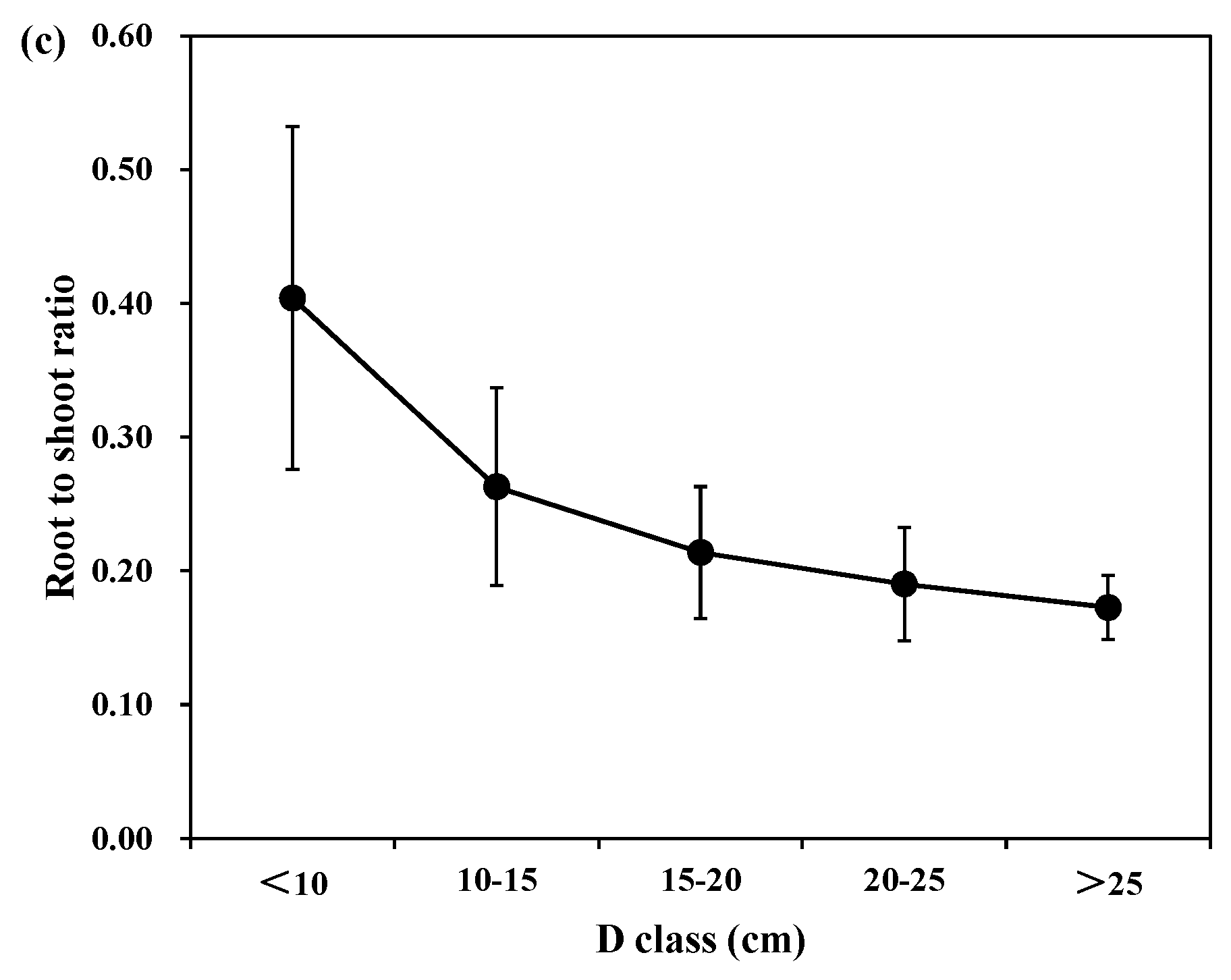
Figure 3 shows the individual root-to -hoot ratio of each tree species in different diameter and age classes. For different diameter classes, the mean root to shoot biomass ratios were 0.35, 0.25, 0.27, 0.28, and 0.27 for Korean pine; 0.21, 0.25, 0.23, 0.25, and 0.29 for larch; and 0.40, 0.26, 0.21, 0.19, and 0.17 for Mongolian pine in D < 10 cm, 10 < D < 15 cm, 15 < D < 20 cm, 20 < D < 25 cm, and D > 25 cm, respectively. Thus, there was a decline in the relative root biomass with the different diameter classes for Korean pine and Mongolian pine, and an increase in the relative root biomass for larch. However, the differences between the medium and large trees were minimal.
3.2. Biomass Additive Systems
With the likelihood analysis, we compared the appropriateness of the two error structures of the two allometric biomass equations (i.e.,
W =
a ×
Db and
W =
a ×
Db ×
Hc) for each of the three species, following the method of Xiao et al. [
31]. The information statistics (ΔAICc) of the likelihood analysis are found in
Table 3. The results indicated that using the LR was favored over NLR to construct the biomass systems (
Table 3). In this study, we developed two additive systems of biomass equations for the three species. The first additive system (i.e., Equation (1), System 1) of biomass equations was fitted in order to estimate individual tree biomass (kg/tree) from tree diameter
D only. The second system (i.e., Equation (2), System 2) enabled the estimation of individual tree biomass from both tree diameter (
D) and total height (
H).
SUR was used to guarantee the additivity property of the tree biomass equations. The independent variable D only or D-H were included in the different biomass component equations (i.e., stems, roots, branches, and foliage), as well as in the total biomass equation. The additivity was guaranteed by setting three constraints to the parameters of the two additive systems of biomass equations in this study.
After applying logarithmic transformation, the coefficient estimates and goodness-of-fit statistics (i.e.,
Ra2 and RMSE) of two additive systems for the three species obtained by SUR are shown in
Table 4. The results indicated that all equations in System 1 (
D only) fitted the biomass data well, with
Ra2 > 0.80 and RMSE < 0.30. The best model fittings were obtained in total, aboveground and stem biomass equations, while the worse model fittings were for foliage and branch biomass equations with relatively lower model
Ra2 and larger RMSE (
Table 4).
When the tree height was measured, both
D and
H were used to develop the second additive system of biomass equations (i.e., Equation (2)). In comparison with the model fitting of Equation (1) (
D only), the second additive system (
D and
H) had a relatively greater
Ra2 and smaller RMSE for total and sub-total component biomass (
Table 4), whereas the stem biomass equation of larch had a greater than 4% increase in
R2 and a greater than 50% decrease in RMSE by including both
D and
H. The SE and
p-values of the parameter estimation indicated that the parameter
c for the predictor
H was statistically significant (at α = 0.05) in eight of the twelve models (
Table 4). Overall, the addition of tree height increased the accuracy for total, aboveground and stem biomass predictions (
Ra2 and RMSE, Equations (3) and (4)), of which the increased range of
Ra2 and RMSE is 0.3%–2% and 0.1%–43% for total biomass of the three species; 0.3%–3% and 8%–49% for aboveground biomass of the three species; and 1%–4% and 18%–50% for stem biomass of the three species, respectively. Adding tree height improved the accuracy for most root, branch, foliage, and crown biomass estimations by no more than 1% in
Ra2 and 10% in RMSE. Among the three species, the additive system with
D or
D-
H for Mongolian pine had slightly better
Ra2 than those of other tree species (
Table 4).
3.3. Model Validation
The model validation statistics (Equations (7)–(10)) based on the jackknifing residuals of the two additive systems of biomass equations are shown in
Table 5, in which the MPE and MPE% represent the average prediction error, and the MAB and MAB% represent the magnitude of prediction error. For all biomass equations of the two additive systems for the three species, the average prediction error, i.e., MPE and MPE%, were close to 0. For total, aboveground and stem biomass of the three species, the magnitude of prediction errors of the two additive systems were relatively small (MAB < 0.2 and MAB% < 7%), and System 2 (Equation (2)) seemed preferable to System 1 (Equation (1)). On the other hand, the biomass equations for root, branch, foliage and crown had less accurate prediction, especially branch and foliage, than those for total, aboveground and stem.
In addition, some researchers suggest the prediction across the diameter classes would be a good way of validating the tree biomass models [
41,
49]. In this study, the MPE, MPE%, MAB and MAB% in tree components of the two additive systems for the three species are listed in
Table A1. Taking the Korean pine as an example, the results indicated that for total biomass, System 2 reduced the prediction error for most diameter classes, but MPE and MPE% increased slightly in the largest diameter class (
D > 25 cm); for aboveground biomass, adding tree height into biomass system reduced the prediction error in all classes; for root biomass, System 2 only reduced the prediction error in
D > 25 cm; for stem biomass, in all diameters System 2 reduced the prediction error; for branch, foliage and crown biomass, System 2 reduced the prediction error in all but the smallest diameter class (
Table A1). The validation of the other plantation species had similar results.
If a log-transformed biomass model is fitted to biomass data, a correction factor (i.e., CF = exp(σ
2/2)) is commonly used to correct for the systematic bias introduced by anti-log transformation. However, Madgwick and Satoo [
50] found that anti-log transformation tended to overestimate biomass by applying the correction factor, and suggested that the correction factor might be ignored if the bias was relatively small compared to the overall variation in the estimate of biomass. In this study, the values of the correction factor (i.e., CF = exp(σ
2/2)) of all biomass equations were less than 1.04, and the percent biases (see [
51], Equation (4)) were relatively small ranging from 0.6% to 4.1% (not shown). Thus, the CF was not applied for the three species in this study.
4. Discussion
We observed a diameter-related pattern in the changes of biomass partitioning among individual aboveground tree components. For larch and Mongolian pine, the results showed that the relative proportion of stem biomass in larger trees was greater across different tree sizes, whereas the crown biomass in larger trees was smaller than those in smaller trees. However, for Korean pine, these findings were not expected and were different from those of other species in general. The main reasons may be (1) the pinecone is an important product of Korean pine. To get more cone production, the older stands of Korean pine need heavier thinning to increase growing space for larger Korean pine trees, which may change the crown profile and the ratio of crown biomass to stems; and (2) the tree climbing and cone taking in the cone collection process may affect tree height growth. Overall, the increase or decrease in the stem, branch, and foliage biomass over different diameter classes found in this study supported the findings of previous studies [
17,
29,
42]. There are obvious differences in partitioning of different biomass components. The belowground biomass (i.e., roots) is a particularly important part of total biomass. The proportion of belowground to total biomass rarely exceeds 30% for most coniferous tree species [
29,
52]. Our results were consistent with the literature of biomass partitioning. The root to shoot biomass ratios found in our study (0.17–0.40) were similar to the range reported for other coniferous species (0.18–0.35) [
29,
52,
53]. It is crucial to accurately estimate the total biomass, and the root to shoot biomass ratios can be an important predictor for root biomass.
Our likelihood analysis showed that data sets of the total, sub-total and components biomass in trees supported a multiplicative error structure so that linear regression on log-transformed data was more appropriate. Our findings were consistent with previous studies that multiplicative error structure is assumed in biomass allometric equations [
4,
11,
29,
51,
54]. The likelihood analysis proposed by several authors [
31,
32] is a good method to verify the error structure of tree biomass data. However, it has not been widely applied in foresty.
Based on the multiplicative error structure of the biomass data, we constructed two additive systems of log-transformed models (Equations (1) and (2)), which were validated using the jackknifing technique. The allometric equation using
D as the only predictor is simple in equation form (i.e., Equation (1)), easy to fit to biomass data, requires only basic forest inventory data to apply in practice, and usually provides reasonably accurate predictions for many species and regions [
11,
14,
19,
55]. However, adding tree height as an additional predictor into biomass equations can significantly improve the model fitting and performance [
21,
28]. Our results demonstrated that adding tree height into the additive system marginally improved two-thirds of the biomass equations for the three plantation species, and were consistent with the literature (e.g., [
20,
26,
28]).
The SUR method should be used in model fitting when total biomass is divided into two parts (aboveground and belowground biomass), aboveground is divided into smaller parts (e.g., stem and crown), crown biomass is divided into two parts (branch and foliage), and stem biomass is divided into bark and wood. The advantages of using an additive system by SUR to estimate total, sub-total and components include: (1) prediction for the components’ biomass sum to the prediction for the total or sub-total biomass; (2) the inherent correlation among biomass components measured on the same sample trees is considered; (3) the parameter estimation is more efficient; and (4) no single biomass component is estimated beyond the total or sub-total biomass [
10,
34,
35]. However, the additivity of biomass equations has not always been addressed when predicting tree total and component biomass.
To date, few biomass equations exist for the three coniferous plantation species of northeastern China, especially Mongolian pine. Wang [
11] developed biomass equations using
D as the only predictor for Korean pine and larch in plantation forest from the Maoershan Ecosystem Research Station of the Northeast Forestry University in Heilongjiang Province, China, which is part of Changbai Mountains. However, his sample size was only ten trees for each species and the biomass equations derived from the sample data were not additive. Our research developed the additive systems of biomass equations for the two species in natural forests in the Xiaoxing’an Mountains, and Changbai Mountains, located in Heilongjiang Province and Jilin Province of China, and the sample sizes in Dong et al. [
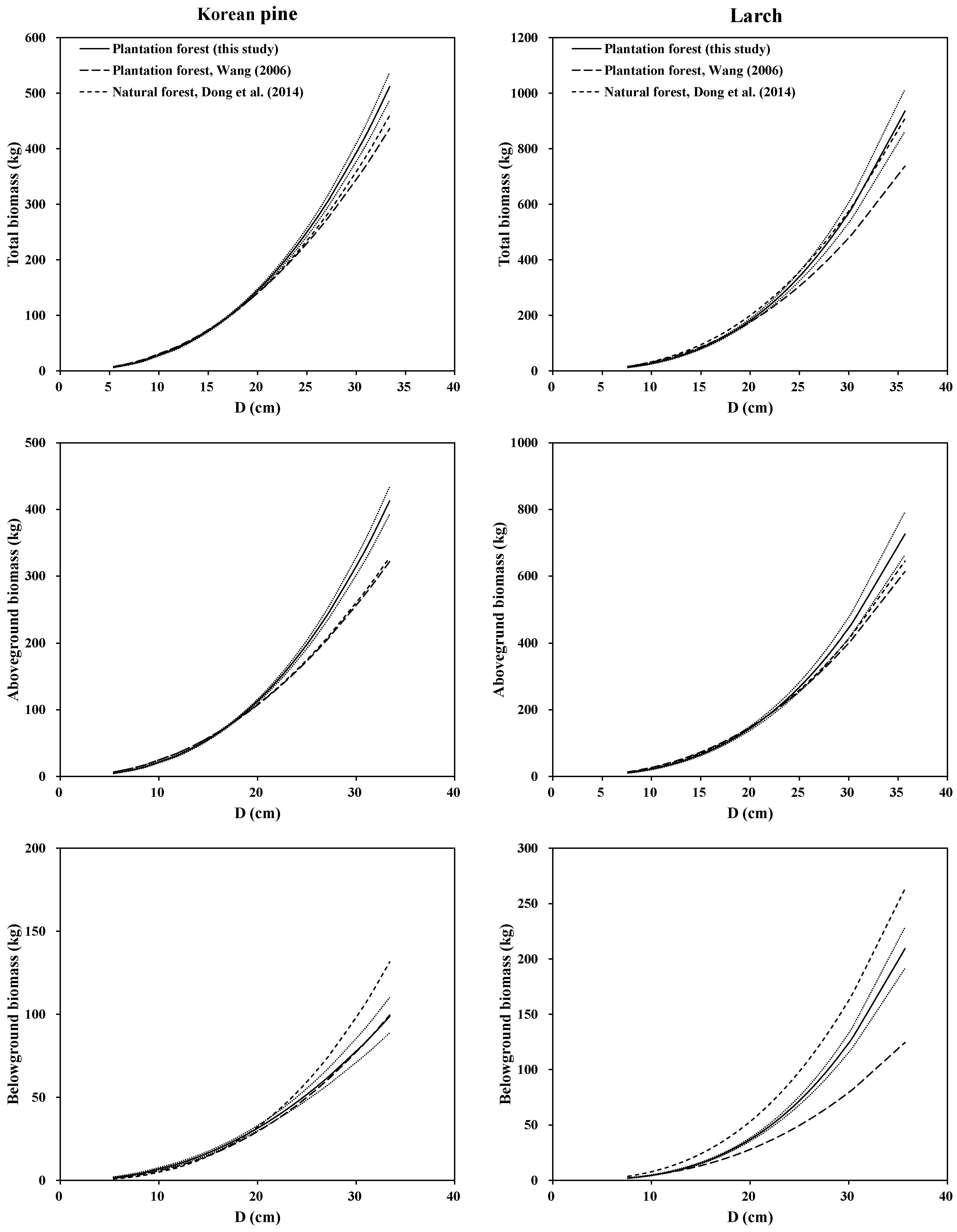
33] was 42 trees for Korean pine and 122 trees for larch in the natural forests, respectively. A graphical analysis of total, aboveground and belowground (root) biomass equations illustrated the differences among our models (System 1) in this study, with Wang [
11]’s biomass equations and Dong et al. [
33]’s biomass equations for the two species (
Figure 4). It is clear that the three models in different forest origins produced different predictions for the root biomass for both species. The models of Korean pine and larch in planted and natural forest produced similar predictions for the aboveground biomass, and most of the mean predicted biomass of Wang’s equations fell into the 95% confidence intervals of mean prediction by our equations. However, our models for the two species in plantations and natural stands yielded higher predictions for the total biomass than Wang’s models for both species, especially for large-sized trees. The possible reasons may be: (1) data of three studies came from different sampling sites; (2) each species of three studies came from different forests; (3) even the two species in plantations were different; and (4) difference in the sample number and sample size range. These may lead to the differences in terms of tree root morphologic features, soil conditions and growth process [
10,
54,
56,
57]. Overall, site, stand origin, and stand structural characteristics of the study trees may also play an important role in biomass estimation and partitioning.
5. Conclusions
In this study, we analyzed the biomass partitioning of aboveground and belowground components across diameter classes for the three coniferous species growing in pure plantations in northeastern P.R. China. Our results were consistent with the literature such that (1) partitioning of aboveground and belowground biomass into various tree components changed considerably with tree diameter; (2) stem biomass for the three species accounted for the largest proportion of total biomass. Among them the relative proportion of stem for larch and Mongolian pine increased with tree diameter, and the relative proportion of foliage and branch decreased, while the proportion patterns of stem, branch and foliage biomass were opposite for Korean pine; and (3) the contribution of root biomass to total tree biomass was rather variable, and the root to shoot ranged from 0.17 to 0.40.
The choice between linear regression on log-transformed data (LR) or nonlinear regression on original data (NLR) depends on the model error structure. We used the likelihood analysis outlined by Xiao et al. [
31] to assess the model error structure of tree biomass equations. The results indicated that the multiplicative error structure was preferable so that logarithmic transformation was necessary. Then two additive biomass systems were developed for the three species. System 1 used tree diameter
D as the only predictor, and System 2 included both tree diameter
D and total height
H as the predictors. As expected, the accuracy of the biomass component equations differed for the two additive systems across the three species. The mean of all models
Ra2 was > 0.94 for System 1 (
D only) and > 0.95 for System 2 (
D and
H). The model root mean square error (RMSE) was relatively smaller for total, aboveground and stem biomass equations, but larger for root, branch, foliage, and crown biomass. Overall, adding tree height into the system of biomass equations can marginally improve model fitting and performance, especially for total, aboveground and stem biomass.