Identification of Nine Pathotype-Specific Genes Conferring Resistance to Fusiform Rust in Loblolly Pine (Pinus taeda L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Host-Pathogen Interaction Matrix
| Host Families (Pine) | Pathogen Isolates (Cqf) | ||||
|---|---|---|---|---|---|
| SC20-21 | NC2-40 | 3327-13-3 | NC2-36 | 0-5-32 | |
| Family 10-5 | I1 | C | C | C | I1 * |
| Family A | C | I2 | C | C | I2 |
| Family 152-329 | C | I3 | I3 | C | I3 |
| Family 29R | C | C | I4 | I4 | C |
| Family B | C5 I9 | C5 C9 | C5 C9 | I5 C9 | C5 I9 |
| Family C | I6 I7 | C6 C7 | I6 C7 | C6 I7 | C6 C7 |
| Family D | I8 | C | C | C | C |
2.2. Pine Families
2.3. Single-Spore/Single-Genotype Isolates and Inoculation Protocol
2.4. Fr Gene Identification
2.5. Fr Gene Mapping
3. Results
3.1. Fr Gene Identification
3.2. Fr Gene Mapping and Marker-Phenotype Analysis of I and C Classifications
3.2.1. Fr1
3.2.2. Fr2
3.2.3. Fr3
3.2.4. Fr4
| Fr1 | Fr2 | Fr3 | Fr4 | Fr5 | Fr6 | Fr7 | Fr8 | Fr9 |
| 10-5 | A | 152-329 | 29R | B | C | C | D | B |
| J7_470+* | BD18_420+ | J7_1840- | 1J7_1840- | 1B20_480- | G13_480- | G13_480+ | 1B20_480+ | J7_1840+ |
| ~1-2 cM | ~7 cM | ~10 cM | ~1.5 cM | ~3 cM | < 6 cM | < 6 cM | ~3 cM | ~ 11 cM |
| AJ4_420- | ACC-CCTA550+ | M18_655+ * | 1F3_1490+ | 1J18_800- | A20_1620- | A20_1620+ | 1A12_725- | K14_440+ |
| ~1-2 cM | ~7 cM | ~13 cM | K3_1530- | 1J18_900+ | < 6 cM | < 6 cM | BD12_1300- | ~13 cM |
| 1BF17_1100- | AY12_1300- | AP12_490- | D20_600- | J7_780+ | J4_565- * | J4_565+ * | AK19_840- | B11_780+ |
| 1BD16_830+ | ~9 cM | W13_915- * | D10_470+ | 2A6_350+ | K3_1530+ | K3_1530- | I10_370 | A9_800+ |
| 1BH1_1300+ | C13_580- | AT15_525- | C4_1750+ | 2D3_1250- * | AP11_415- | AP11_415+ | AJ4_420- | |
| AY14_410+ | AK6_850- | N9_320- | G13_480- | 2AK19_840- | K14_440- | K14_440+ | G13_480- | |
| 2D16_1180+ | 1E12_1200+ | 1H20_400- | A6_1650- | J7_1840- | J7_1840+ | |||
| 2J4_565+* | 1ACG-CCTA550- | 1I10_515+ | A19_580+ | |||||
| F13_780- | ACC-CCGA315+ | 1A12_420+ | ||||||
| J4_1000- | BB11_780+ | D11_1350+ * | ||||||
| 3C12_1050+ | AZ18_850+ | |||||||
| 3C12_450- | ACG-CCAA258- | |||||||
| BF4_1100- | ACG-CCAA532+ | |||||||
| A9_625+ | ||||||||
| D11_1350-* | ||||||||
| H20_400+ | ||||||||
| H20_800- | ||||||||
| A19_500+ | ||||||||
| K1_900- | ||||||||
| B8_650- |
3.2.5. Fr5
3.2.6. Fr6 and Fr7
3.2.7. Fr8
3.2.8. Fr9
3.3. Genomic Localization of Fr Genes
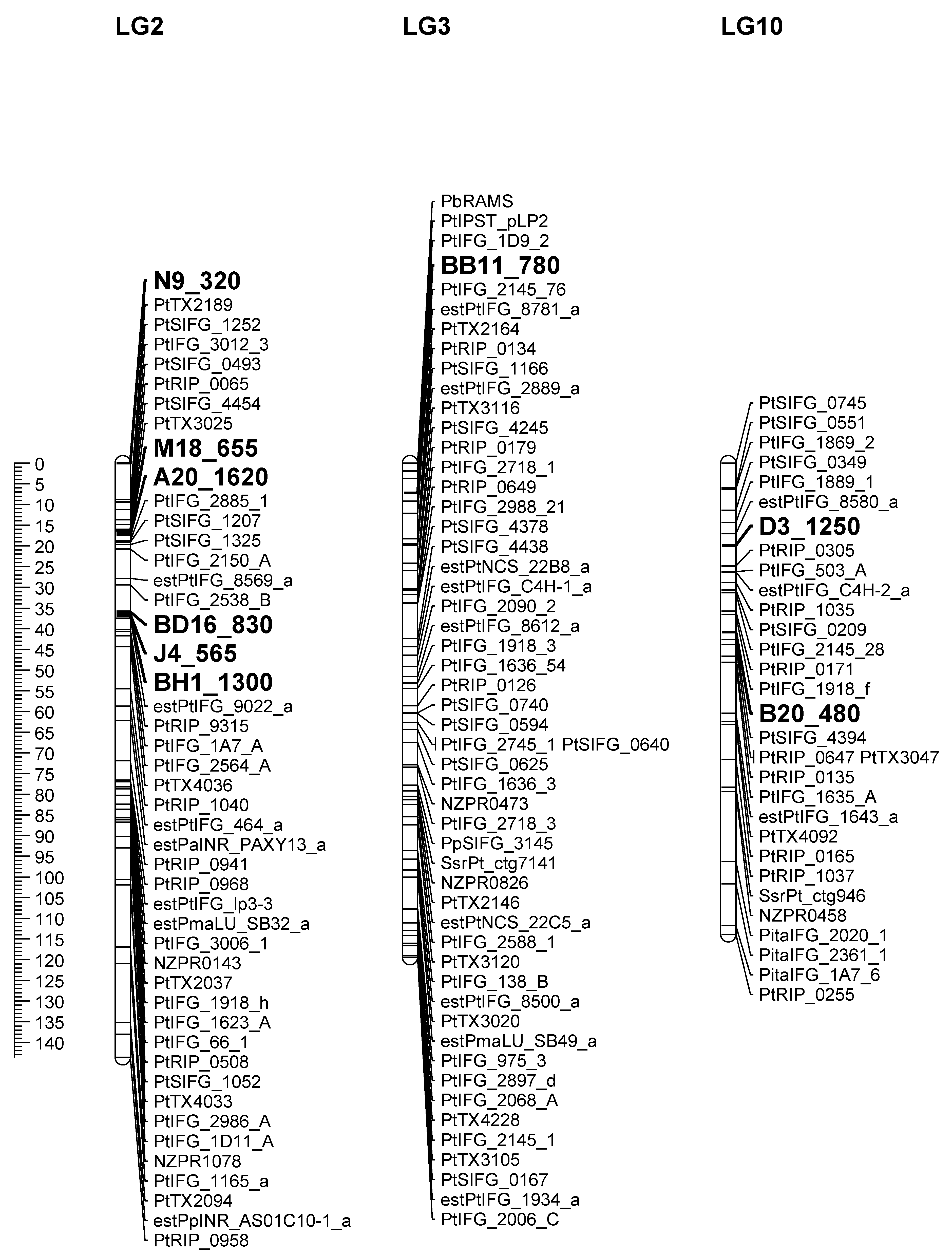
| RAPD Markers | Fr Gene/Cluster | Pedigree | Reference LG |
|---|---|---|---|
| BD16_830 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | base | 2 |
| BH1_1300 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | base | 2 |
| J4_565 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | base | 2 |
| N9_320 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | base | 2 |
| BB11_780 | Fr2 | base | 3 |
| B20_480 | Fr5, Fr8 | base | 10 |
| A20_1620 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | qtl | 2 |
| BD16_830 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | qtl | 2 |
| BH1_1300 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | qtl | 2 |
| M18_655 | Fr1, Fr3, Fr4, Fr6, Fr7, Fr9 | qtl | 2 |
| BB11_780 | Fr2 | qtl | 3 |
| D3_1250 | Fr5, Fr8 | qtl | 10 |
4. Discussion
4.1. Fr Gene Identification and Genomic Mapping
4.2. Fr2 Clarification
4.3. Summation and Perspectives
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Powers, H.R., Jr.; McClure, J.P.; Knight, H.A.; Dutrow, G.F. Incidence and financial impacts of fusiform rust in the south. J. For. 1974, 72, 398–401. [Google Scholar]
- Anderson, R.L.; McClure, J.P.; Cost, N.C.; Uhler, R.L. Estimating fusiform rust losses in five southeastern states. South. J. Appl. For. 1986, 10, 237–240. [Google Scholar]
- Cubbage, F.; Pye, J.; Holmes, T.; Wagner, J. An economic analysis of fusiform rust protection research. South. J. Appl. For. 2000, 24, 77–85. [Google Scholar]
- Barber, J.C.; Dorman, K.W.; Bauer, E. Slash Pine Progeny Tests Indicate Genetic Variation in Resistance to Rust; Research Notes, Number 104, U.S. Department of Agriculture Forest Service Southeastern Forest Experiment Station: Asheville, NC, USA, 1957. [Google Scholar]
- Powers, H.R., Jr.; Kraus, J.F. Developing fusiform rust-resistant loblolly and slash pines. Plant Dis. 1983, 67, 187–189. [Google Scholar] [CrossRef]
- Snow, G.A.; Matthews, F.R.; Nance, W.L.; Foster, G.S. Effects of pollen source on loblolly pine resistance to Cronartium quercuum f. sp. fusiforme. For. Sci. 1990, 36, 304–312. [Google Scholar]
- McKeand, S.E.; Li, B.; Amerson, H.V. Genetic variation in fusiform rust resistance in loblolly pine across a wide geographic range. Silvae Genet. 1999, 48, 255–260. [Google Scholar]
- Matthews, F.R.; Rowan, J. An improved method for large-scale inoculations of pine and oak with Cronartium fusiforme. Plant Dis. Rep. 1972, 56, 931–934. [Google Scholar]
- Snow, G.A.; Kais, A.G. Technique for inoculating pine seedlings with Cronartium fusiforme. In Biology of Rusts of Forest Trees; Bingham, R.T., Hoff, R., McDonald, G.T., Eds.; Miscellaneous Publication 1221, U.S. Department Agriculture Forest Service: Washington, DC, USA, 1972; pp. 325–326. [Google Scholar]
- Powers, H.R., Jr.; Schmidt, R.A.; Snow, G.A. Current status and management of fusiform rust on southern pines. Ann. Rev. Phytopathol. 1981, 19, 353–371. [Google Scholar] [CrossRef]
- Zobel, B.; Talbert, J. Applied Forest Tree Improvement; John Wiley and Sons: New York, NY, USA, 1984. [Google Scholar]
- Jewell, F.F. Inheritance of rust resistance in southern pines. In Breeding Pest-Resistant Trees; Gerhold, H.D., Schreiner, E.J., McDermott, R.E., Winieski, J.A., Eds.; Pergamon Press: Oxford, UK, 1966; pp. 107–109. [Google Scholar]
- Jewell, F.F.; Mallett, S.L. Testing slash pine for rust resistance. For. Sci. 1967, 13, 413–418. [Google Scholar]
- Kinloch, B.B., Jr.; Walkinshaw, C.H. Resistance to fusiform rust in southern pines: How is it inherited? In Rusts of Pine; Hiratsuka, Y., Samoil, J.K., Blenis, P.V., Crane, P.E., Laishley, B.L., Eds.; Information Report, NOR-X-317, Northern Forestry Centre, Forestry Canada: Edmonton, AB, Canada, 1991; pp. 219–228. [Google Scholar]
- Griggs, M.M.; Walkinshaw, C.H. Diallel analysis of genetic resistance to Cronartium quercuum f. sp. fusiforme in slash pine. Phytopathology 1982, 72, 816–818. [Google Scholar] [CrossRef]
- Powers, H.R., Jr. Pathogenic variation among single-aeciospore isolates of Cronartium quercuum f.sp. fusiforme. For. Sci. 1980, 26, 280–282. [Google Scholar]
- Kuhlman, E.G.; Matthews, F.R. Variation in virulence among single-aeciospore isolates from single-gall isolates of Cronartium quercuum f. sp. fusiforme. Can. J. For. Res. 1993, 23, 67–71. [Google Scholar] [CrossRef]
- Nelson, C.D.; Doudrick, R.L.; Nance, W.L.; Hamaker, J.M.; Capo, B. Specificity of host-pathogen genetic interaction for fusiform rust disease on slash pine. In Proceedings of the 22nd Southern Forest Tree Improvement Conference, Atlanta, GA, USA, 14–17 June 1993; pp. 403–411. Available online: http://www.rngr.net/publications/tree-improvement-proceedings/sftic/1993/specificity-of-host-pathogen-genetic-interaction-for-fusiform-rust-disease-on-slash-pine (accessed on 30 July 2015).
- Wilcox, P.L.; Amerson, H.V.; Kuhlman, E.G.; Lui, B.H.; O’Malley, D.M.; Sederoff, R. Detection of a major gene for resistance to fusiform rust disease in loblolly pine by genomic mapping. Proc. Natl. Acad. Sci. USA 1996, 93, 3859–3864. [Google Scholar] [CrossRef] [PubMed]
- Amerson, H.V.; Jordan, A.P.; Kuhlman, E.G.; O’Malley, D.M.; Sederoff, R.R. Genetic basis of fusiform rust disease resistance in loblolly pine. In Proceedings of the 24th Southern Forest Tree Improvement Conference, Orlando, FL, USA, 9–12 June 1997; p. 403. Available online: http://www.rngr.net/publications/tree-improvement-proceedings/sftic/1997/genetic-basis-of-fusiform-rust-disease-resistance-in-loblolly-pine (accessed on 30 July 2015).
- Jordan, A.P. Fusiform Rust Disease Resistance and Genomic Mapping in Loblolly Pine. Master’s Thesis, Department of Forestry, North Carolina State University, Raleigh, NC, USA, 1997. [Google Scholar]
- Kuhlman, E.G.; Amerson, H.V.; Jordan, A.P.; Pepper, W.D. Inoculum density and expression of major gene resistance to fusiform rust disease in loblolly pine. Plant Dis. 1997, 81, 597–600. [Google Scholar] [CrossRef]
- Stelzer, H.E.; Doudrick, R.L.; Kubisiak, T.L.; Nelson, C.D. Prescreening slash pine and Cronartium pedigrees for evaluation of complementary gene action in fusiform rust disease. Plant Dis. 1999, 83, 385–389. [Google Scholar] [CrossRef]
- Amerson, H.V.; Kubisiak, T.L.; Garcia, S.A.; Kuhlman, E.G.; Nelson, C.D.; McKeand, S.E.; Mullin, T.J.; Li, B. Interacting genes in the pine-fusiform rust forest pathosystem. In Proceedings of the 28th Southern Forest Tree Improvement Conference, Raleigh, NC, USA, 21–23 June 2005; p. 60. Available online: http://www.rngr.net/publications/tree-improvement-proceedings/sftic/2005/interacting-genes-in-the-pine-fusiform-rust-forest-pathosystem (accessed on 30 July 2015).
- Kubisiak, T.L.; Amerson, H.V.; Nelson, C.D. Genetic interaction of the fusiform rust fungus with resistance gene Fr1 in loblolly pine. Phytopathology 2005, 95, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.D.; Amerson, H.V.; Kubisiak, T.L. Unravelling and managing fusiform rust disease: A model approach for coevolved forest tree pathosystems. For. Pathol. 2010, 40, 67–72. [Google Scholar] [CrossRef]
- Kubisiak, T.L.; Anderson, C.L.; Amerson, H.V.; Smith, J.A.; Davis, J.M.; Nelson, C.D. A genomic map enriched for markers linked to Avr1 in Cronartium quercuum f.sp. fusiforme. Fungal Genet. Biol. 2011, 48, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Kubisiak, T.L.; Roberds, J.H.; Spaine, P.C.; Doudrick, R.L. Microsatellite DNA suggests regional structure in the fusiform rust pathogen Cronartium quercuum f.sp. fusiforme. Heredity 2004, 92, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Burdine, C.S.; Kubisiak, T.L.; Johnson, G.; Nelson, C.D. Fifty-two polymorphic microsatellite loci in the rust fungus, Cronartium quercuum f.sp. fusiforme. Mol. Ecol. Notes 2007, 7, 1005–1008. [Google Scholar] [CrossRef]
- Kolmer, J. Leaf rust of wheat: Pathogen biology, variation and host resistance. Forests 2013, 4, 70–84. [Google Scholar] [CrossRef]
- Myburg, H.; Morse, A.M.; Amerson, H.V.; Kubisiak, T.L.; Huber, D.; Osborne, J.A.; Garcia, S.A.; Nelson, C.D.; Davis, J.M.; Covert, S.F.; et al. Differential gene expression in loblolly pine (Pinus taeda L) challenged with the fusiform rust fungus, Cronartium quercuum f.sp. fusiforme. Physiol. Mol. Plant Pathol. 2006, 68, 79–91. [Google Scholar] [CrossRef]
- Remington, D.L.; Whetten, R.W.; Liu, B.-H.; O’Malley, D.M. Construction of an AFLP genetic linkage map with nearly complete genome coverage in Pinus taeda. Theor. Appl. Genet. 1999, 98, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Myburg, A.A.; Remington, D.L.; O’Malley, D.M.; Sederoff, R.R.; Whetten, R.W. High throughput AFLP analysis using infrared dye-labeled primers and an automated DNA sequencer. BioTechniques 2001, 30, 348–357. [Google Scholar] [PubMed]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Shmoys, D.B.; Durrett, R.T.; Tanksley, S.D. Selective mapping: A strategy for optimizing the construction of high-density linkage maps. Genetics 2000, 55, 401–420. [Google Scholar]
- Echt, C.S.; Saha, S.; Krutovsky, K.V.; Wimalanathan, K.; Erpelding, J.E.; Liang, C.; Nelson, C.D. An annotated genetic map of loblolly pine based on microsatellite and cDNA markers. BMC Genet. 2011, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Li, H.; Ghosh, S.; Amerson, H.; Li, B. Major gene detection for fusiform rust resistance using Bayesian complex segregation analysis in loblolly pine. Theor. Appl. Genet. 2006, 113, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, P.L. Genetic Dissection of Fusiform Rust Resistance in Loblolly Pine. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 1995. [Google Scholar]
- Li, H. Major Gene Detection for Fusiform Rust Resistance Using Segregation Analysis and Linkage Analysis in Loblolly Pine. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2003. [Google Scholar]
- Isik, F.; Amerson, H.V.; Whetten, R.W.; Garcia, S.A.; McKeand, S.E. Interactions of Fr genes and mixed-pathogen inocula in the loblolly pine-fusiform rust pathosystem. Tree Genet. Genomes 2012, 8, 15–25. [Google Scholar] [CrossRef]
- Quesada, T.; Resende, M.F.R., Jr.; Munoz, P.; Wegrzyn, J.L.; Neale, D.B.; Kirst, M.; Peter, G.F.; Gezan, S.A.; Nelson, C.D.; Davis, J.M. Mapping fusiform rust resistance genes within a complex mating design of loblolly pine. Forests 2014, 5, 347–362. [Google Scholar] [CrossRef]
- Neale, D.B.; Wegrzyn, J.L.; Stevens, K.A.; Zimin, A.V.; Puiu, D.; Crepeau, M.W.; Cardeno, C.; Koriabine, M.; Holtz-Morris, A.E.; Liechty, J.D.; et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Amerson, H.V.; Wilcox, P.; Sederoff, R.R.; Kuhlman, E.G.; O’Malley, D.M.; Grattapaglia, D. Methods for Within Family Selection of Disease Resistance in Woody Perennials Using Genetic Markers. U.S. Patent #5,908,978, 1 June 1999. [Google Scholar]
- Kong, X. RAPD Mapping and Its Application in Slash Pine Breeding. Ph.D. Thesis, Texas A&M University, College Station, TX, USA, 1996. [Google Scholar]
- Millar, C.L.; Kinloch, B.B. Taxonomy, phylogeny, and coevolution of pines and their stem rusts. In Rusts of Pine; Hiratsuka, Y., Samoil, J.K., Blenis, P.V., Crane, P.E., Laishley, B.L., Eds.; Information Report, NOR-X-317, Northern Forestry Centre, Forestry Canada: Edmonton, AB, Canada, 1991; pp. 1–38. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amerson, H.V.; Nelson, C.D.; Kubisiak, T.L.; Kuhlman, E.G.; Garcia, S.A. Identification of Nine Pathotype-Specific Genes Conferring Resistance to Fusiform Rust in Loblolly Pine (Pinus taeda L.). Forests 2015, 6, 2739-2761. https://doi.org/10.3390/f6082739
Amerson HV, Nelson CD, Kubisiak TL, Kuhlman EG, Garcia SA. Identification of Nine Pathotype-Specific Genes Conferring Resistance to Fusiform Rust in Loblolly Pine (Pinus taeda L.). Forests. 2015; 6(8):2739-2761. https://doi.org/10.3390/f6082739
Chicago/Turabian StyleAmerson, Henry V., C. Dana Nelson, Thomas L. Kubisiak, E. George Kuhlman, and Saul A. Garcia. 2015. "Identification of Nine Pathotype-Specific Genes Conferring Resistance to Fusiform Rust in Loblolly Pine (Pinus taeda L.)" Forests 6, no. 8: 2739-2761. https://doi.org/10.3390/f6082739






