1. Introduction
Accurate forest biomass mapping methods are required globally and could be utilized e.g., for predicting forest aboveground biomass (AGB), bioenergy potential and carbon stock. The use of forest-based bioenergy is also increasing and will play a major role in the bioenergy sector in the near future. At the moment, a typical approach to mapping AGB over large areas is based on generalizing field sample plot measurements using coarse- or medium-resolution remote sensing (RS) data and other numeric map data. For sample plots, the amount of AGB is estimated using local allometric models (e.g., [
1,
2]). This approach, referred to as Multi-Source National Forest Inventory (MS-NFI), was first introduced and tested in Finland in the early 1990s (e.g., [
3]) and is nowadays routinely used in the Nordic countries to generate biomass maps for various purposes [
3].
Forest management practices, e.g., in Finland, are based on intensive, small-scale forestry, which results in mosaic-like forest structures and stand sizes of approximately 2 ha on average. Mapping AGB over larger areas under such land use or vegetation patterns is challenging based on medium- or coarse-resolution RS data with pixel sizes varying from 30 m × 30 m (e.g., Landsat 7) to 250 m × 250 m (e.g., MODIS). Previous studies have shown that the mismatch between field measurement data, an individual pixel of satellite data [
4] and the scarcity of available field data are the main challenges in forest attribute prediction. Coarse pixel size in the RS data results in mixed pixels on stand boundaries and thus an inaccurate description of the vegetation structure variability especially when looking at relatively small areas (e.g., forest stands of 1–3 hectares or field-measured sample plots). For example, Tuominen
et al. [
5] used Landsat 5 TM imagery with a resampled pixel size of 25 m × 25 m to predict total volume and AGB. Estimated root-mean-squared error (RMSE) of the total volume and AGB varied between 65.1% and 73.1% of the mean plot-level reference values. Such accuracy levels are not acceptable for detailed AGB mapping, which is required e.g., in bioenergy harvesting. More detailed methods are therefore needed for AGB mapping, especially at the local (plot or pixel) level.
Airborne laser scanning (ALS) is a three-dimensional (3D) measurement technique, which is becoming increasingly popular in forest resource mapping. ALS data can be used to predict a variety of forest inventory attributes with a high level of accuracy [
6,
7] due to its capability of describing 3D vegetation structure. ALS has two main approaches for estimating forest information: an area-based approach (ABA, [
6]) and individual tree detection (ITD, [
8]). Forest AGB prediction with laser scanning techniques has previously been studied at varying levels, from single trees (e.g., [
9,
10,
11,
12,
13,
14]) to larger areas. Recent overviews are provided by Zolkos
et al. [
15] and Popescu and Hauglin [
16].
The coefficient of determination values (
R2) between predicted AGB and laser-derived metrics have previously varied between 0.32 and 0.92 depending on, e.g., tree species and development classes. Combined small-footprint ALS and multispectral airborne data were used in Popescu
et al. [
17] to predict AGB in deciduous and pine (
Pinus L.) forests at a resolution of 168 m
2. The maximum
R2 values were 0.32 for deciduous and 0.82 for pine dominated sample plots. Aardt
et al. [
18] improved the
R2 values for deciduous trees to 0.58 using ALS point height metrics as predictors in a per-segment prediction of AGB. The highest
R2 values were reported by Næsset [
19], who used regression methods to predict AGB for 143 sample plots (size of 300–400 m
2) in young and mature coniferous forests. The regression models explained 92% of the AGB variability for all forest types.
Latifi
et al. [
20] compared different nearest neighbor (NN) and random forest (RF) techniques for the prediction of volume and AGB in southwestern Germany, with the target areas for which the AGB values were predicted varying from 13 m
2 to 452 m
2. They concluded that the RF method was superior to other NN methods. ABA accuracy depends highly on the coverage of different forest structures within the training data and the prediction technique used [
7]. According to Fassnacht
et al. [
21] the data type (RS) had the highest impact on the AGB prediction accuracy. Prediction technique was seen as the second most important factor. Fassnacht
et al. [
21] also concluded that increasing the sample size of the modeling data did not effectively improve the AGB prediction accuracy. Stepper
et al. [
22] confirmed this conclusion and reported that decreasing the sample size from 1875 plots to 226 plots had a minimal effect on the model performance. Traditionally the required training data for the ABA is measured in the field, which is costly, especially in remote areas. Vastaranta
et al. [
23] used ITD to replace field measurements required for the ABA. This method was further tested for AGB prediction in Kankare
et al. [
24]. The AGB prediction RMSEs varied between 23.0 and 32.3 tons per ha. Tuominen and Haapanen [
25] reported similar or slightly higher prediction accuracies using a combination of ALS and digital aerial photograph-derived metrics.
The National Land Survey of Finland (NLS) began collecting ALS data over the entire area of Finland in 2008 to provide a new national-level elevation model. The data are available free of charge from their data services. As corresponding data are also increasingly available in several other countries, e.g., Sweden, Italy, Spain, USA, Netherlands and England, (e.g., [
26,
27,
28]), these data are expected to have a great potential and a wide variety of possible applications in forestry and especially in forest biomass mapping and monitoring. Although the data are essentially acquired for ground elevation modeling, Nord-Larsen and Schumacher [
29] and Villikka
et al. [
30] verified the suitability of these data also for forest attribute prediction. However, they did not compare the obtained accuracies with any existing inventories such as the MS-NFI. To better justify bringing a new method to operational forestry, it should be shown to yield at least at a similar level of accuracy as the presently used inventory methods. Overall, no studies reporting component biomass prediction accuracies based on sparse density, leaf-off ALS data currently exist in the literature.
The main objective of our study was thus to evaluate the accuracy of predicting AGB and its components (stem, living branch and canopy biomass) using sparse density, leaf-off ALS data collected essentially for elevation modeling. The ALS metrics were derived at plot-level (300 m2) and tested as predictor variables to predict AGB components. Results were compared to the Multi-Source National Forest Inventory (MS-NFI, ©Finnish Forest Research Institute 2012) AGB estimates. Similar datasets are being collected in several countries worldwide, and the comparison is therefore seen as important towards detailed large-scale forest resource mapping applications. Since the ALS data were acquired with multiple sensors and varying scanning parameters, we also derived the corresponding metrics from a canopy height model (CHM) and tested this type of feature extraction as an alternative strategy to eliminate the sensor effects. Our detailed objectives were: (1) to evaluate the accuracy of the AGB component prediction at a resolution of 300 m2; (2) to compare AGB prediction accuracy to an existing approach of predicting AGB over large areas; and (3) to evaluate the accuracy and effects of the CHM derived metrics on the AGB component prediction accuracy.
4. Discussion
The low density, leaf-off ALS data collected by NLS have a great potential in more detailed forest mapping and monitoring applications compared to the mapping applications that are currently widely utilized. The main two benefits in the use of these types of data are the free availability and full coverage of Finland in the near future. Similar data sets are increasingly available from several other countries worldwide. Challenges, on the other hand, include: (1) the low pulse densities and leaf-off acquisitions of the data; (2) potentially multiple sensors and varying data acquisition parameters in operational setups; and (3) requirements for local, acquisition-specific field reference data for training. Few previous studies have demonstrated the suitability of these data types for forest attribute predictions, but introducing a new operational forest mapping method requires the accuracy to be at least at a similar level and costs should be reduced compared to existing inventories (e.g., MS-NFI), in which the forest attribute predictions are readily available without a separate modeling step. The main objective of our study was therefore to evaluate the prediction accuracy of AGB and its components (stem, living branch and canopy biomass), using metrics derived from sparse density leaf-off ALS data, essentially acquired for elevation modeling, as predictor variables and to compare the prediction accuracies to MS-NFI AGB estimates in Finland. Our study also evaluated the accuracy and effect of the conventional ALS metrics computed from the CHM to equalize the differences in the point clouds recorded caused by multiple sensors and varying scanning parameters.
ALS is one of the most promising remote sensing techniques for forest resource mapping applications and its implementation to operational forestry began a few years ago. ALS data acquisition will be challenging in practice because the rapid development of the scanning technologies means that the datasets will be collected with varying sensors and scanning parameters (e.g., NLSs ALS data). ALS data collected with multiple sensors and varying data acquisition parameters is shown to have an effect on forest attribute prediction (see e.g., [
37]). Differences between ALS sensors will cause dissimilarity in point cloud properties, which will affect the metrics derived from the point clouds [
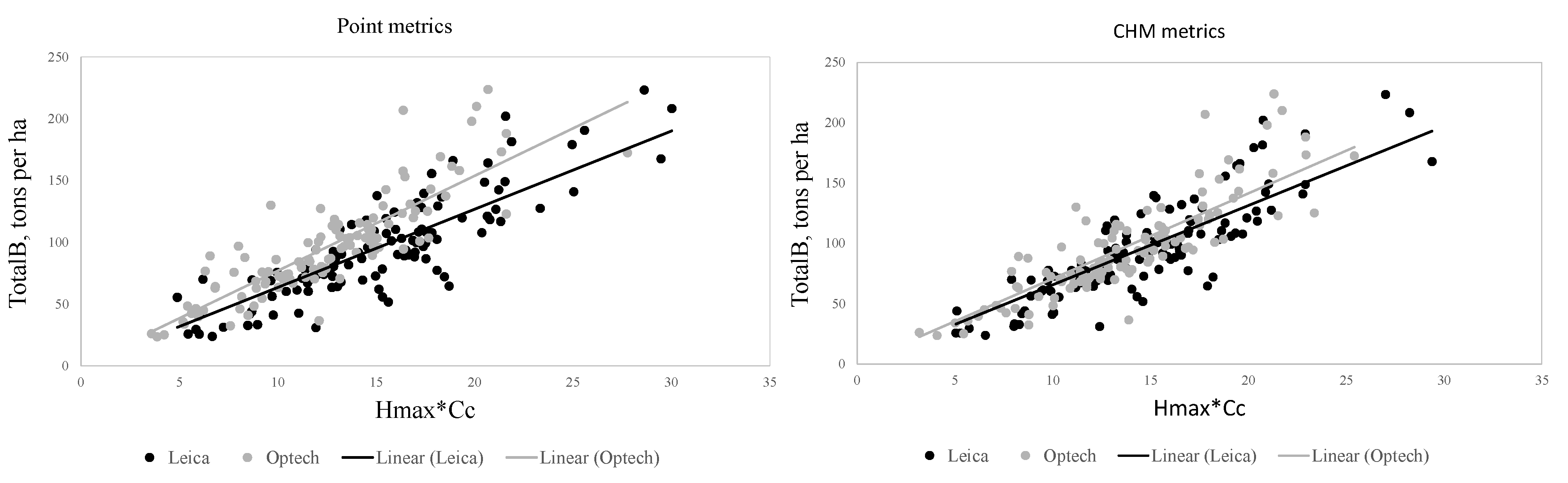
37]. This effect needs to be minimized if the data are to be used for large-scale mapping applications. The ALS data sets used in our study were collected with two different scanning sensors, as described in
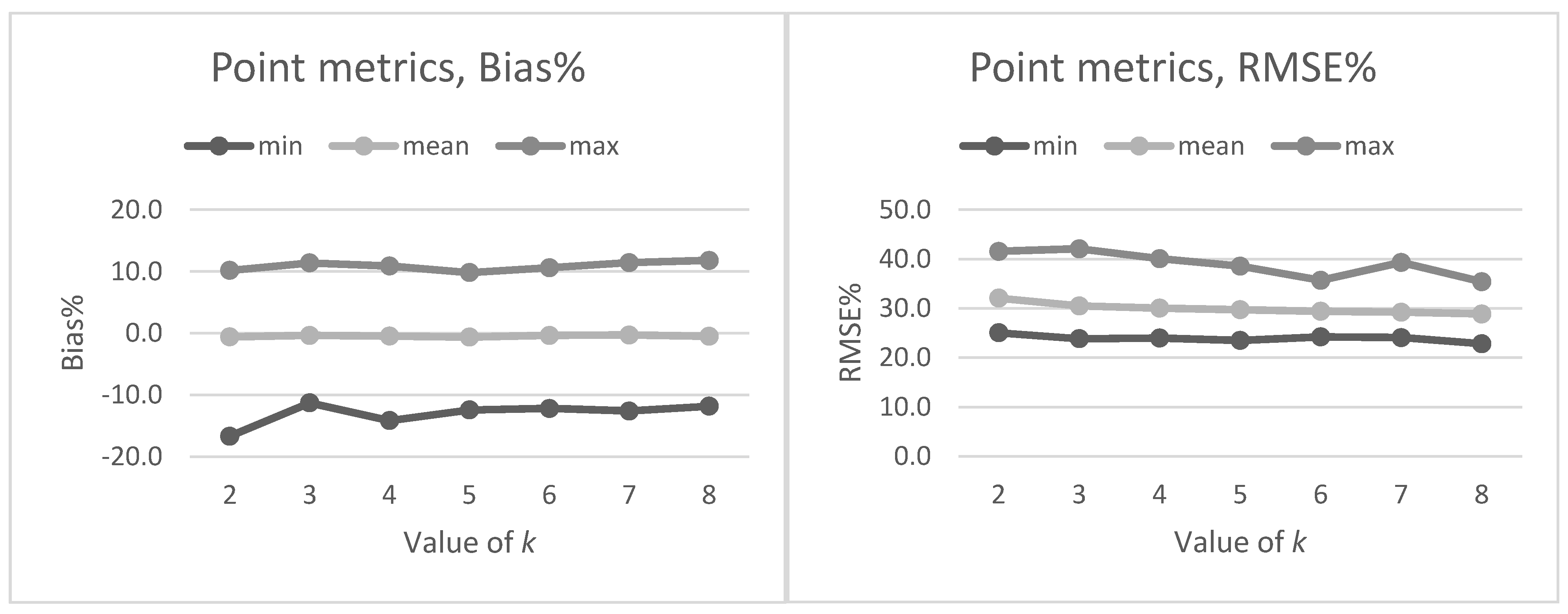
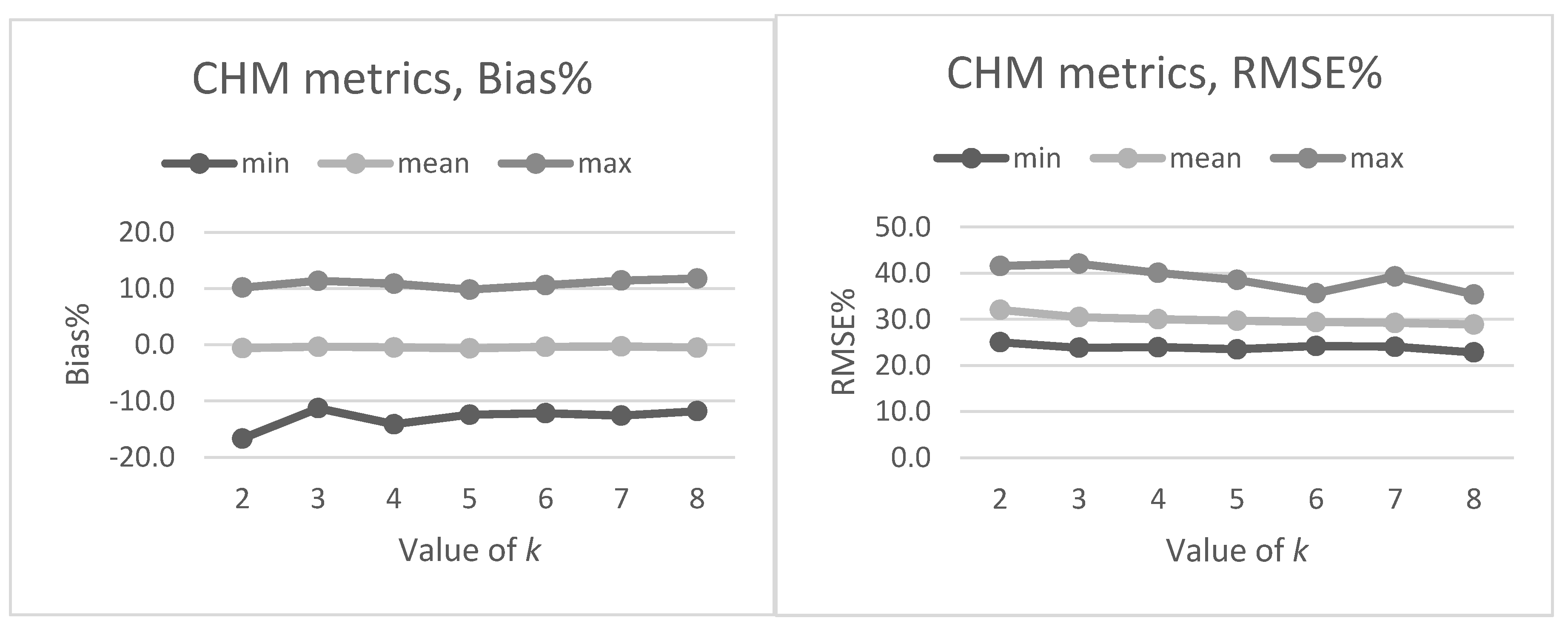
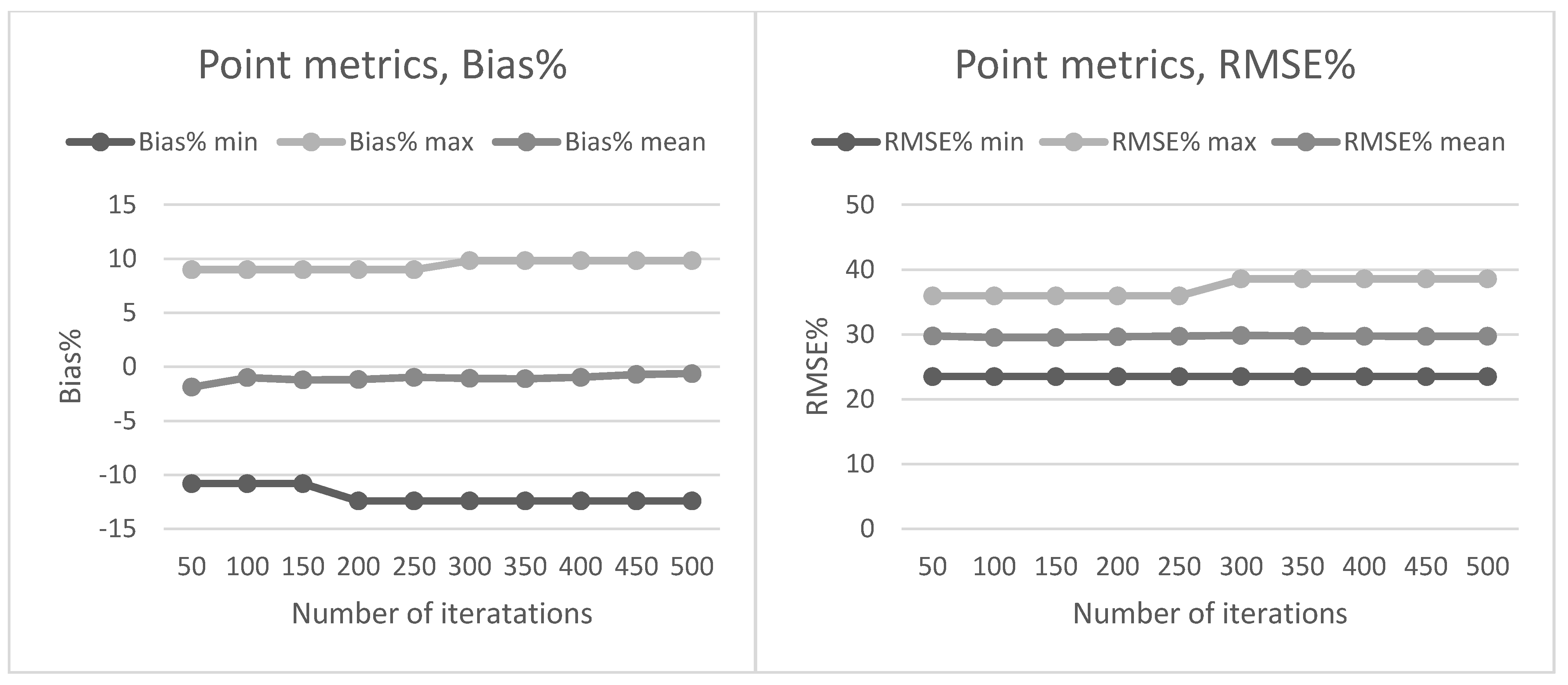
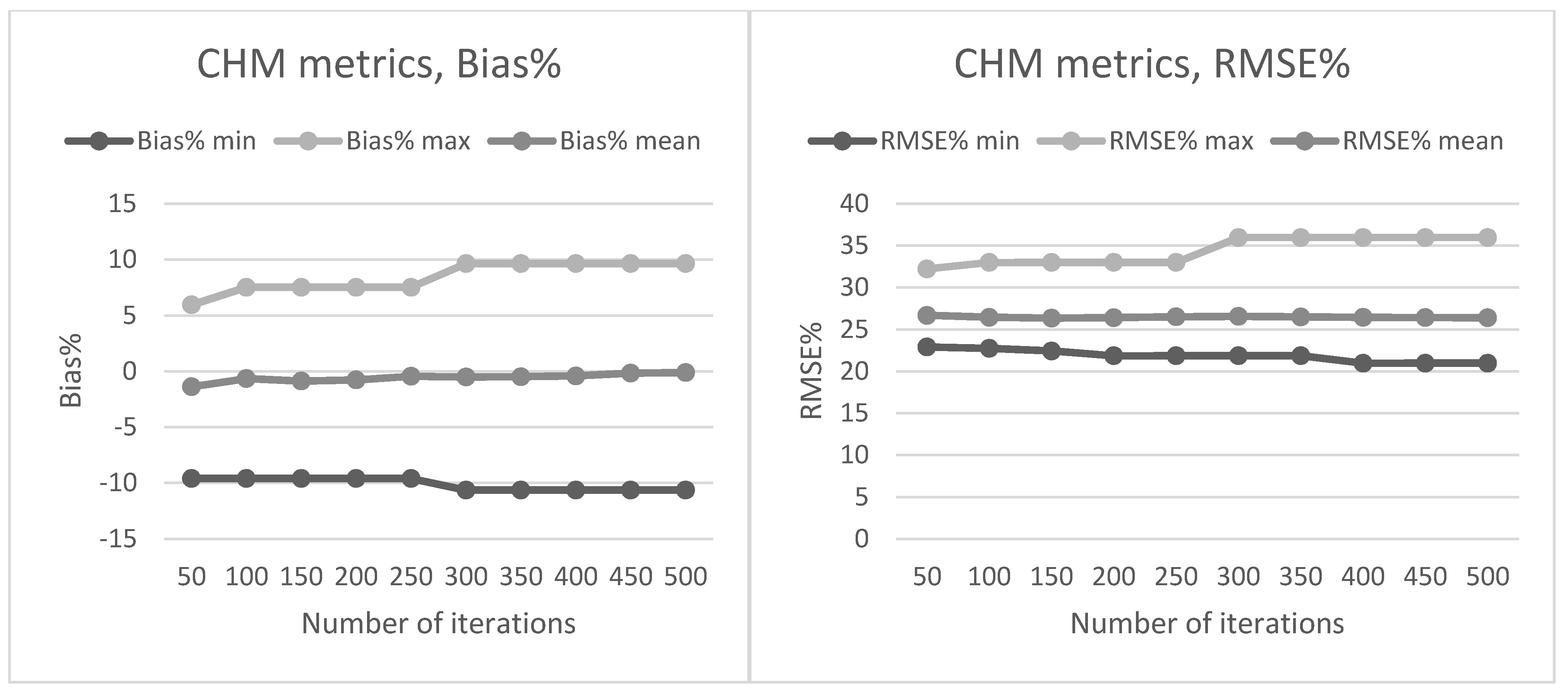
Section 2.2. The preliminary tests in our present study also showed that data acquisition parameters and sensors affected the metrics derived for predictions and thus the forest attribute predictions. The estimated bias caused by this effect was successfully reduced using the CHM-derived metrics as predictor variables in the NN predictions. The interpolation of the CHM simplifies the metrics used because it utilizes only the highest echo inside each pixel, but it still provides a detailed representation of the forest structure in terms of the variation of the top surface height. The effect was further evaluated by repeating the NN predictions 500 times to assess the model performance in detail. The CHM-derived metrics were slightly more stable and resulted in a smaller estimated bias but the difference was not as clear as in the first results. Therefore, this approach warrants further testing where all the external factors (such as data composition, area coverage and forest structure) are minimized.
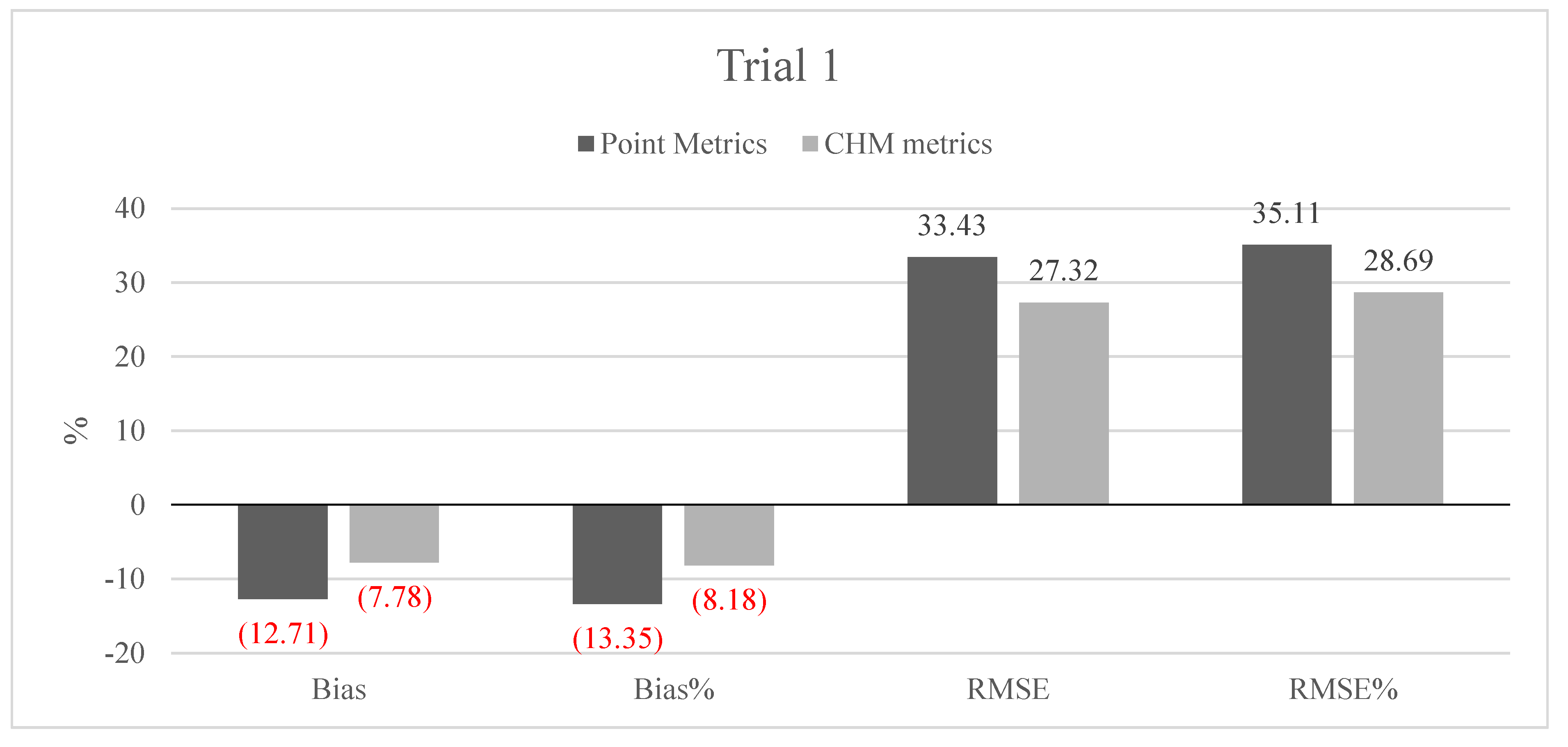
The AGB components were predicted with similar accuracies to that reported previously (e.g., [
17,
24,
25,
38]) using the NN approach with both metric data sets (point or CHM). The RMSE of totalB was 29.7% using ALS point metrics as predictor variables. The accuracy was further improved using CHM-derived metrics. The RMSE decreased to 26.4% and the minor negative estimated bias visible with the point metrics was reduced to near zero. Kankare
et al. [
24] used higher-density ALS data in the same study area (Evo), and achieved an RMSE of 24.9% for totalB. Latifi
et al. [
20] achieved plot-level accuracies of 22.2%–45.5% for totalB, using high-density ALS data. The best results were achieved with the RF approach in comparison to other NN methodologies [
20]. Vastaranta
et al. [
39] compared ALS and high spatial resolution digital stereo imagery (DSI) in forest attribute prediction and achieved RMSE% of 17.5% and 23.7%, respectively. The results presented in our present study were achieved at a much lower pulse density (0.8 points per m
2) than Kankare
et al. [
24], Latifi
et al. [
20] or Vastaranta
et al. [
39], which shows the potential of this data set in forest mapping applications. The use of the DSI-derived metrics provides another cost-efficient option for forest AGB mapping and monitoring [
39]. However, to adequately compare the results and methods used, all external factors (e.g., site-specific or a selection of model features) should be removed.
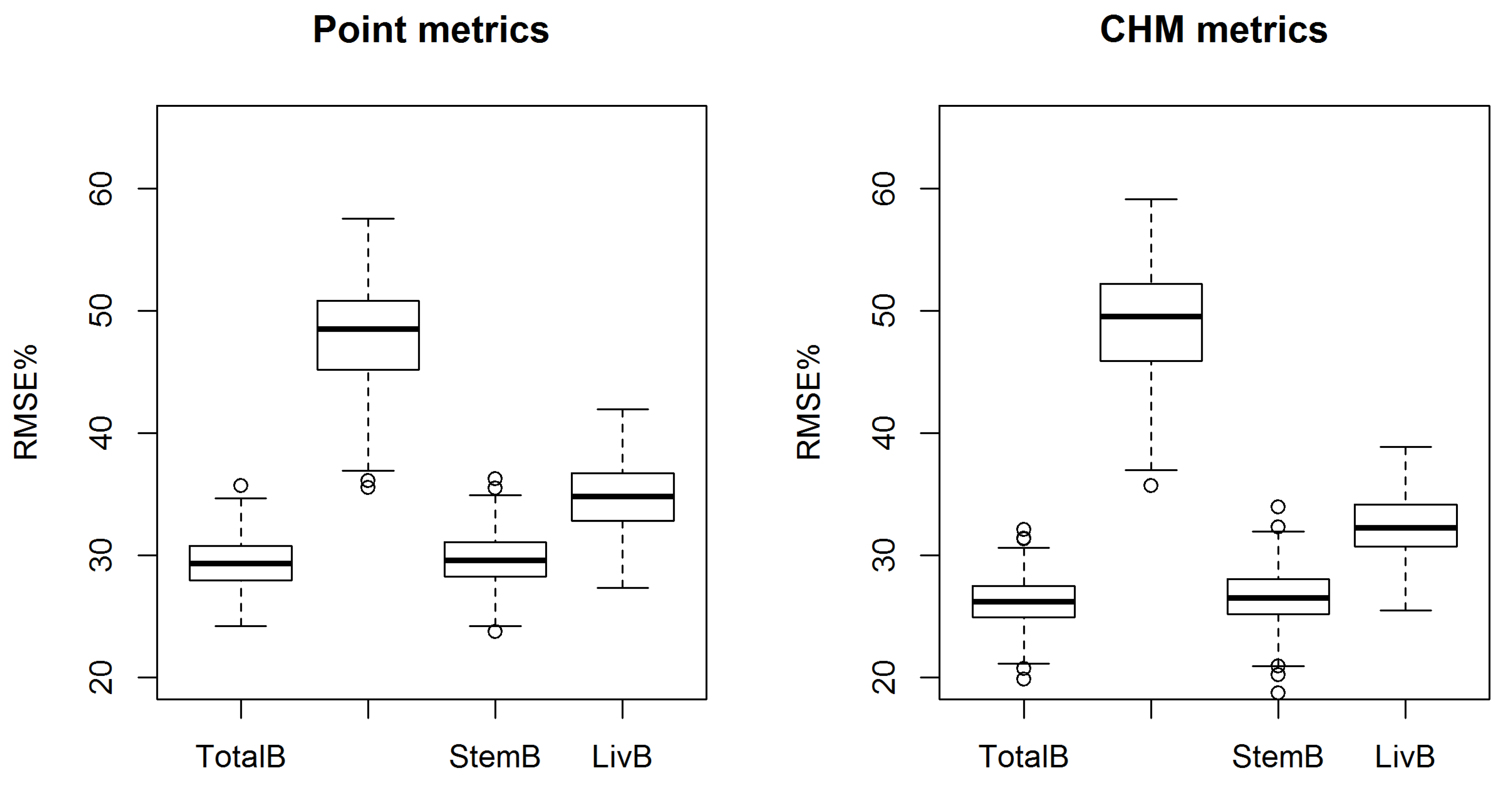
The AGB component prediction accuracies were also improved using the CHM metrics. Only the RMSE of canopyB was increased (by 1.3%), but the decrease in the estimated bias was notable. The increase in the RMSE was most probably caused by the amount of dead branch biomass, which is shown to be a challenging attribute to predict [
24]. He
et al. [
40] reported even higher accuracies for component biomass prediction using ALS data with similar densities. The ABA methods are also highly dependent on the training data available and in our study we were able to use 126 circular field-measured sample plots (~300 m
2) in training. The increment of accuracy in He
et al. [
40] compared to our present study is most likely caused by the difference in sample plot size and the difference in forest structure and conditions. He
et al. [
40] used either 20 m × 20 m or 25 m × 25 m sample plots, an increase which tends to improve the prediction accuracies (e.g., [
41,
42]). Optimization of the amount and plot size of required training data for forest mapping with low density ALS data would be an important topic for future research.
The data acquisition period was not optimized for forest attribute prediction, and it is one important development steps for the method used here. The leaf-off period can especially affect the prediction accuracy of deciduous tree-dominated forests, but in our study area the deciduous trees are a minority and coniferous trees (Scots pine and Norway spruce) are the dominant tree species, comprising 78.2% of the total volume.
The sparse density, leaf-off ALS-data are an interesting possible alternative for cost-efficient forest mapping over larger areas. Nord-Larsen and Schumacher [
29] and Villikka
et al. [
30] previously verified that this type of data collected for elevation modeling could also be suitable for forest attribute prediction, but did not compare the accuracy levels obtained with any existing technique such as MS-NFI. To be plausibly considered by the actors of operational forestry, a new forest-mapping method should have an accuracy comparable to that produced by existing inventory methods. The accuracies achieved in our study are higher, compared to those of the MS-NFI with the RMSE of totalB 47.7%, which was notably lower than the results based on the ALS data in our study. The difference in the prediction accuracy was caused by the prediction methods and the RS data used. MS-NFI utilizes satellite images and NFI-measured sample plots, resulting in 20 m × 20 m pixel-sized thematic maps on the forest attributes. The resolution of the RS data and the resampling procedure will have an effect on the prediction accuracies, which could potentially be minimized with higher resolution RS data. The data resolution used in our study was 1 m × 1 m for the DTM and CHM. The location accuracy (
x,
y) of the sample plots will have an effect on the prediction accuracies, but it is minimized during the field measurement campaigns by positioning the sample plot completely inside the forest stand. The location accuracy will have the most effect if the sample plot is located at the border of forest stand or in the middle of two stands.