Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population Sampling
2.2. DNA Extraction and Amplification
2.3. Data Analysis
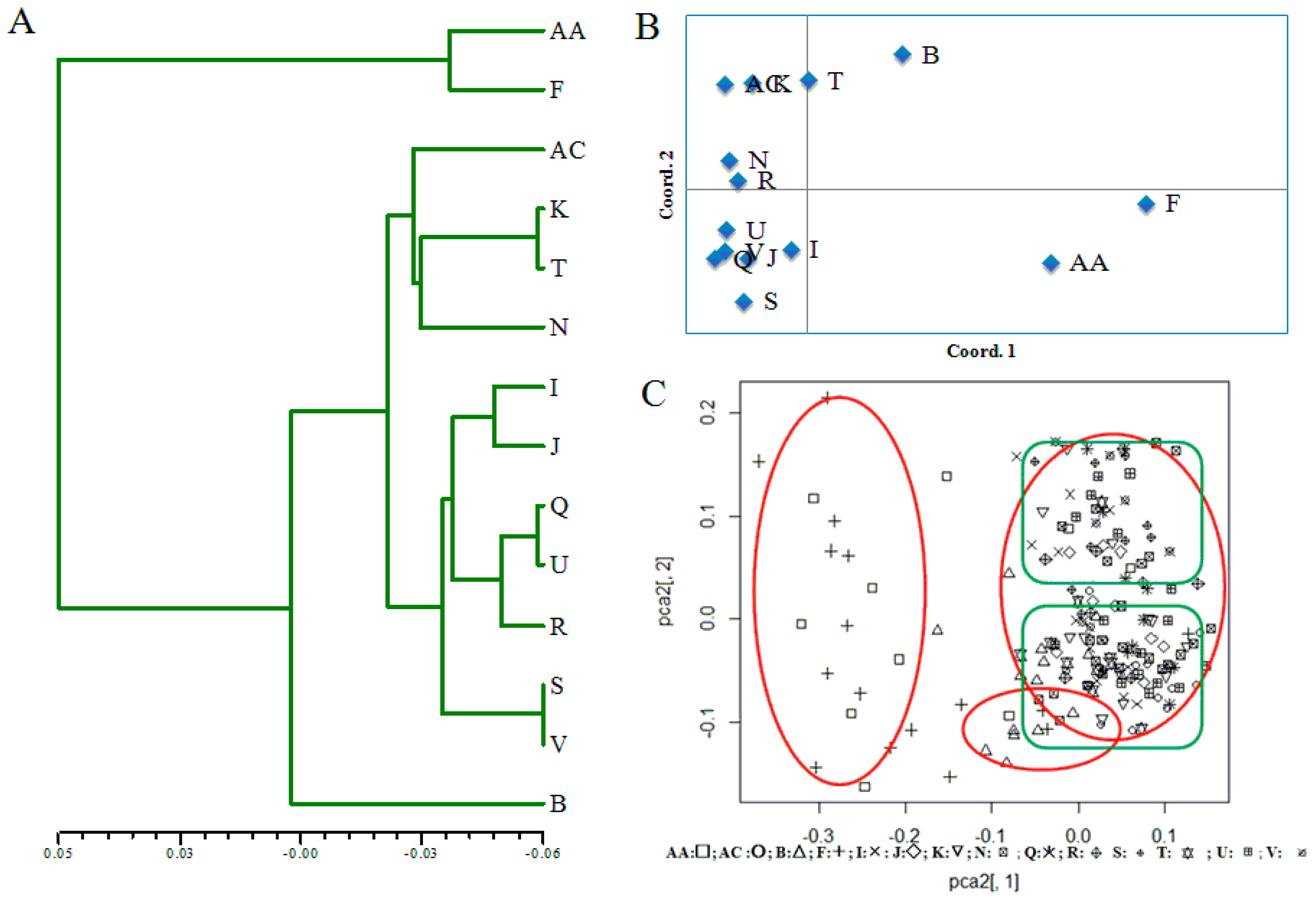
3. Results
3.1. Population Genetic Diversity
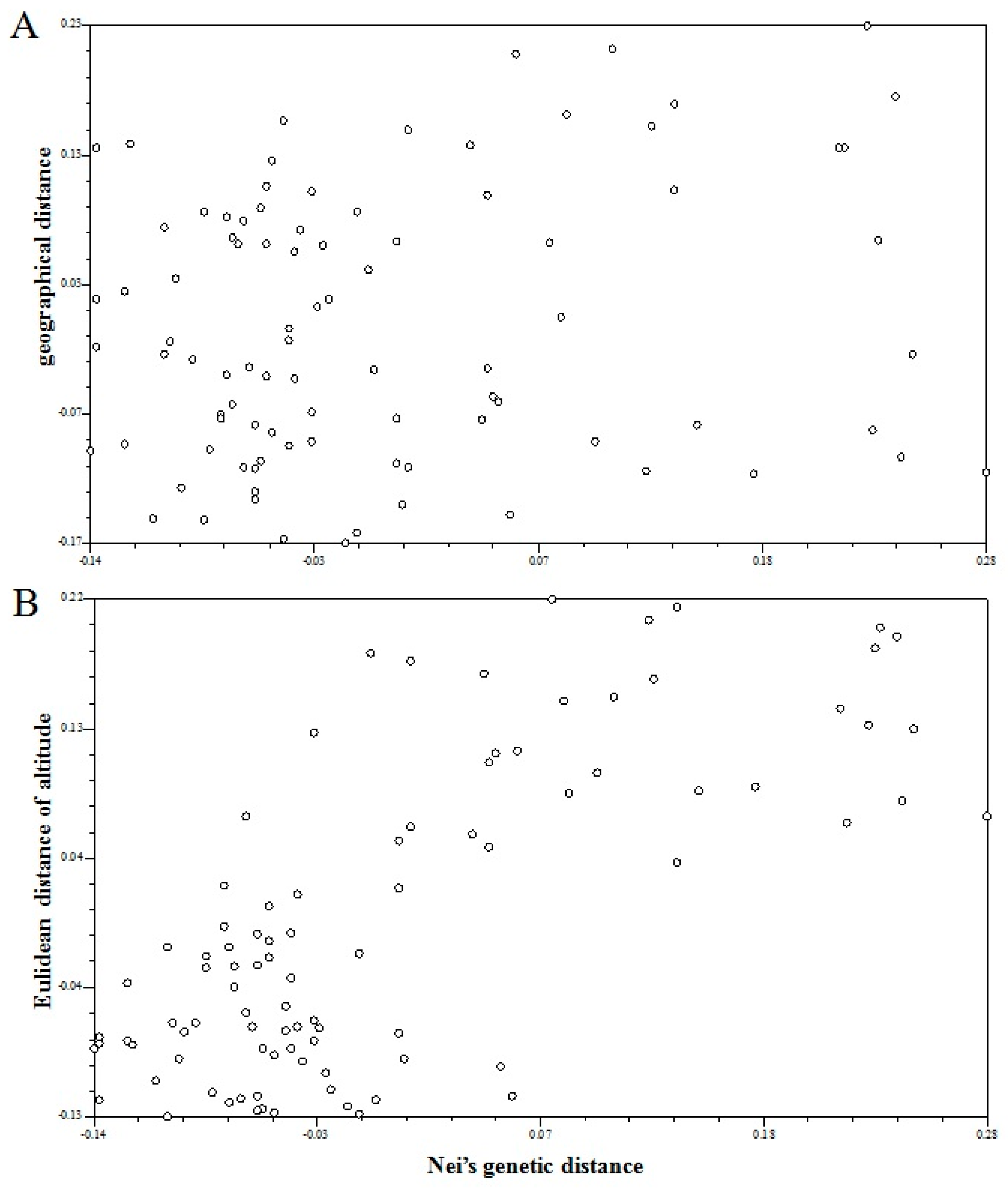
3.2. Population Genetic Differentiation
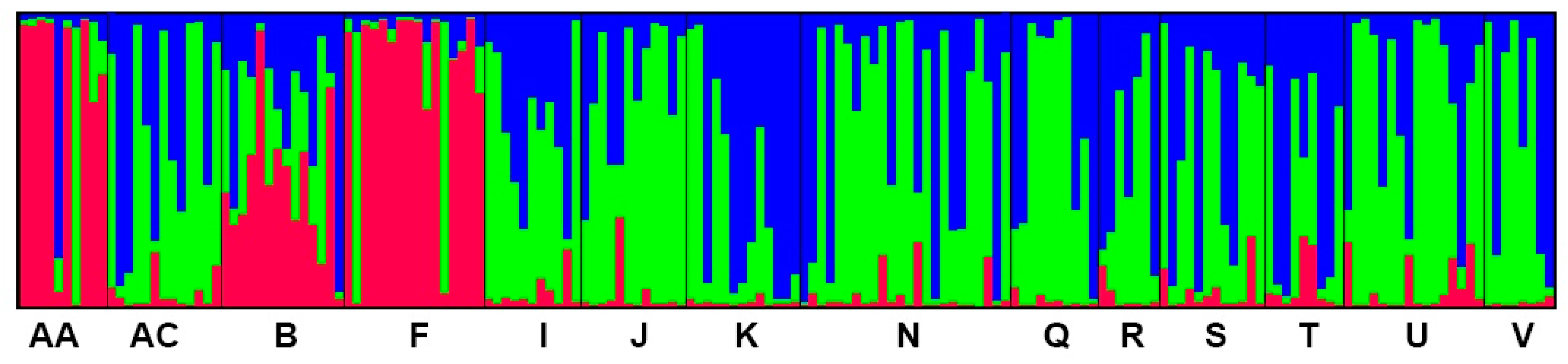
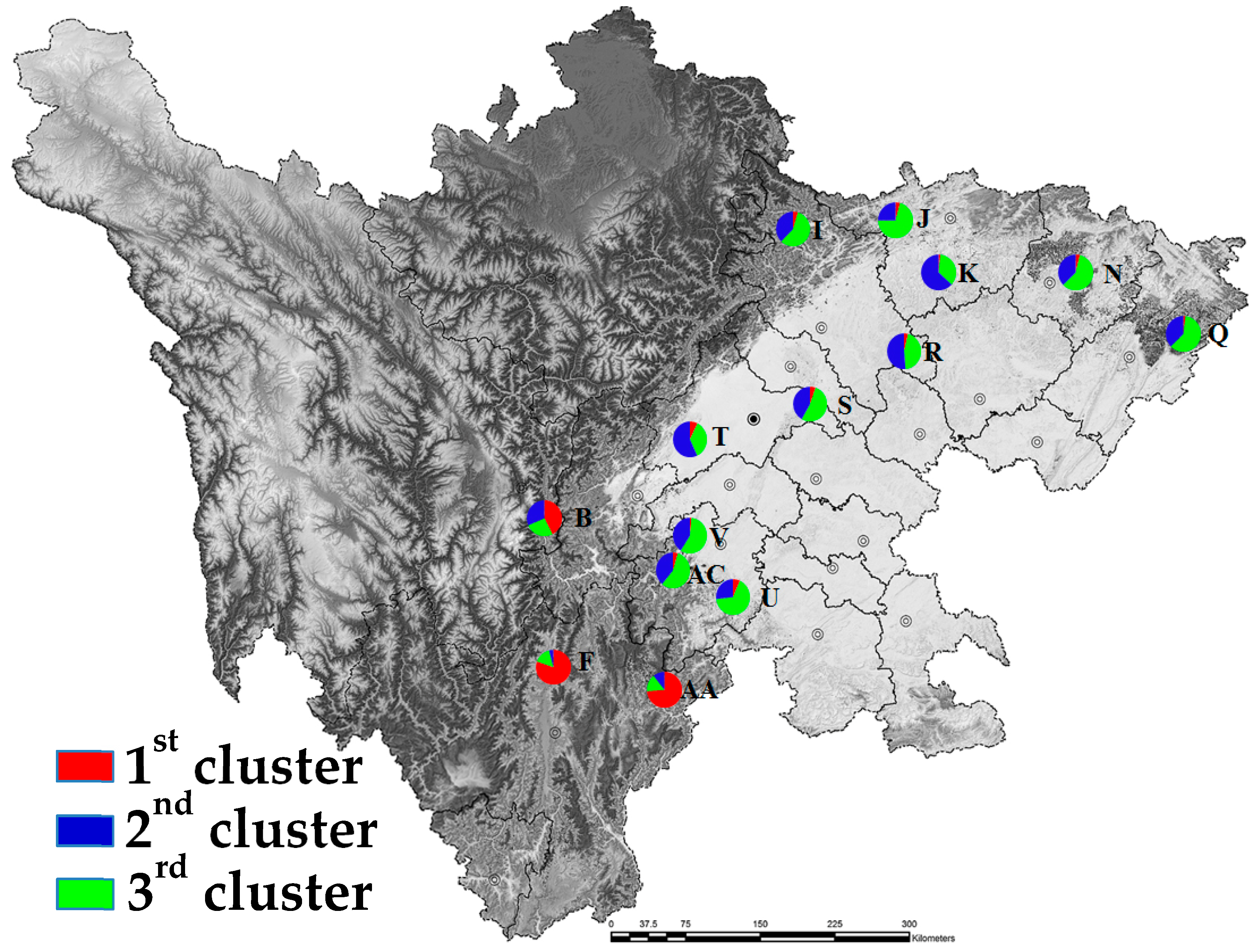
3.3. Population Genetic Structure
4. Discussion
4.1. Polyploidy and Data Analysis in A. cremastogyne
4.2. Population Genetic Structure and Geographical Variation in A. cremastogyne
4.3. Genetic Improvement of A. cremastogyne
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, Z.D. Phylogeny and phytogeography of the Betulaceae (cont.). Acta Phytotax. Sin. 1994, 32, 101–153. (In Chinese) [Google Scholar]
- Kuang, K.; Li, P.; Cheng, S.; Lu, A. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1979; Volume 21, pp. 93–103. (In Chinese) [Google Scholar]
- Chen, M.; Chen, J.; Wu, J.; Cheng, S. Variation of fruiting quantity and nut and seed characters of Alnus cremastogyne Clones. Sci. Silvae Sin. 2008, 44, 153–156. (In Chinese) [Google Scholar]
- Gu, Y.; Wang, Q.; Luo, J. A study on the phenotypic diversity of fruits of natural populations of Alnus cremastogyne Burk in Sichuan. J. Sichuan For. Sci. Technol. 2009, 30, 19–22. (In Chinese) [Google Scholar]
- Chen, Y.; Li, G.; Wang, H. Study on phenotypic variation in natural range of longpeduncled alder (Alnus cremastogyne). For. Res. 1999, 12, 379–385. (In Chinese) [Google Scholar]
- Wang, J. Study on Genetic Variation and Selection of Alnus cremastogyne; Beijing Forestry University: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Zhuo, R.; Chen, Y. Genetic variation of different Alnus cremastogyne populations. J. Zhejiang For. Sci. Technol. 2005, 25, 13–16. (In Chinese) [Google Scholar]
- Rao, L.; Yang, H.; Guo, H.; Duan, H.; Chen, Y. Development of SSR molecular markers based on transcriptome sequences of Alnus. For. Res. 2016, 29, 875–882. (In Chinese) [Google Scholar]
- Zhuk, A.; Veinberga, I.; Daugavietis, M.; Ruņģis, D. Cross-species amplification of Betula pendula Roth. simple sequence repeat markers in Alnus Species. Bal. For. 2018, 14, 116–121. [Google Scholar]
- Lance, S.L.; Jones, K.L.; Hagen, C.; Glenn, T.C.; Jones, J.M.; Gibson, J.P. Development and characterization of nineteen polymorphic microsatellite loci from seaside alder, Alnus maritima. Conserv. Genet. 2009, 10, 1907–1910. [Google Scholar] [CrossRef]
- Jones, J.M.; Gibson, J.P. Population genetic diversity and structure within and among disjunct populations of Alnus maritima (seaside alder) using microsatellites. Conserv. Genet. 2011, 12, 1003–1013. [Google Scholar] [CrossRef]
- Jones, J.M.; Gibson, J.P. Mating system analysis of Alnus maritima (seaside alder), a rare riparian tree. Castanea 2012, 77, 11–20. [Google Scholar] [CrossRef]
- Lepais, O.; Muller, S.D.; Ben Saad-Limam, S.; Benslama, M.; Rhazi, L.; Belouahem-Abed, D.; Daoud-Bouattour, A.; Gammar, A.M.; Ghrabi-Gammar, Z.; Bacles, C.F. High genetic diversity and distinctiveness of rear-edge climate relicts maintained by ancient tetraploidisation for Alnus glutinosa. PLoS ONE 2013, 8, e75029. [Google Scholar] [CrossRef] [PubMed]
- Havrdova, A.; Douda, J.; Krak, K.; Vit, P.; Hadincova, V.; Zakravsky, P.; Mandak, B. Higher genetic diversity in recolonized areas than in refugia of Alnus glutinosa triggered by continent-wide lineage admixture. Mol. Ecol. 2015, 24, 4759–4777. [Google Scholar] [CrossRef]
- Mingeot, D.; Husson, C.; Mertens, P.; Watillon, B.; Bertin, P.; Druart, P. Genetic diversity and genetic structure of black alder (Alnus glutinosa [L.] Gaertn) in the Belgium-Luxembourg-France cross-border area. Tree Gene. Genom. 2016, 12, 1–12. [Google Scholar] [CrossRef]
- Mandak, B.; Havrdova, A.; Krak, K.; Hadincova, V.; Vit, P.; Zakravsky, P.; Douda, J. Recent similarity in distribution ranges does not mean a similar postglacial history: A phylogeographical study of the boreal tree species Alnus incana based on microsatellite and chloroplast DNA variation. New. Phytol. 2016, 210, 1395–1407. [Google Scholar] [CrossRef]
- Cubry, P.; Gallagher, E.; O’Connor, E.; Kelleher, C. Phylogeography and population genetics of black alder (Alnus glutinosa (L.) Gaertn.) in Ireland: Putting it in a European context. Tree Gene. Genomes 2015, 11, 99. [Google Scholar] [CrossRef]
- Mingeot, D.; Baleux, R.; Watillon, B. Characterization of microsatellite markers for black alder (Alnus glutinosa [L.] Gaertn). Conserv. Genet. Res. 2010, 2, 269–271. [Google Scholar] [CrossRef]
- Lepais, O.; Bacles, C.F.E. De Novo discovery and multiplexed amplification of microsatellite markers for black alder (Alnus glutinosa) and related species using SSR-enriched shotgun pyrosequencing. J. Hered. 2011, 102, 627–632. [Google Scholar] [CrossRef]
- Yang, A.; Wu, B.; Shen, C.; Zhang, G.; Liu, H.; Guo, H.; Li, J.; Liu, J.; Zhou, L.; Rao, L.; et al. Microsatellite records for volume 9, issue 3. Conserv. Genet. Res. 2017, 9, 507–511. [Google Scholar] [CrossRef]
- Clark, L.V.; Jasieniuk, M. Polysat: An R package for polyploid microsatellite analysis. Mol. Ecol. Res. 2011, 11, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G.; Van Tienderen, P.H. Genotype and genodive: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- Slatkin, M.; Barton, N.H. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 1989, 43, 1349–1368. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Rohlf, F.J. NTSYS-pc numerical taxonomy and multivariate analysis system. Am. Stat. 1987, 41, 330. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [PubMed]
- Mandak, B.; Vit, P.; Krak, K.; Travnicek, P.; Havrdova, A.; Hadincova, V.; Zakravsky, P.; Jarolimova, V.; Bacles, C.F.; Douda, J. Flow cytometry, microsatellites and niche models reveal the origins and geographical structure of Alnus glutinosa populations in Europe. Ann. Bot. 2016, 117, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Vít, P.; Douda, J.; Krak, K.; Havrdová, A.; Mandák, B. Two new polyploid species closely related to Alnusglutinosa in Europe and North Africa—An analysis based on morphometry, karyology, flow cytometry and microsatellites. Taxon 2017, 66, 567–583. [Google Scholar] [CrossRef]
- Ren, B.; Liu, J. Cytological study on Alnus in China(I). Guihaia 2006, 26, 356–359. (In Chinese) [Google Scholar]
- Yang, H.; Rao, L.; Guo, H.; Duan, H.; Chen, Y. Karyotyping of five species of Alnus in east Aisa region. J. Plant Genet. Res. 2013, 14, 203–207. (In Chinese) [Google Scholar]
- Hong, D. Plant Cytotaxonomy; Science Press: Beijing, China, 1990. (In Chinese) [Google Scholar]
- Murai, S. Phytotaxonomical and geobotanical studies on genus Alnus in Japan (III). Taxonomy of whole world species and distribution of each section. Bull. Gov. For. Exp. Stat. 1964, 171, 1–107. [Google Scholar]
- Dering, M.; Latałowa, M.; Boratyńska, K.; Kosiński, P.; Boratyński, A. Could clonality contribute to the northern survival of grey alder [Alnus incana (L.) Moench] during the Last Glacial Maximum? Acta Soc. Bot. Pol. 2016, 86, 3523. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Population, Vol. 4. Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; pp. 97–159. [Google Scholar]
- Su, T.; Spicer, R.A.; Li, S.-H.; Xu, H.; Huang, J.; Sherlock, S.; Huang, Y.-J.; Li, S.-F.; Wang, L.; Jia, L.-B.; et al. Uplift, climate and biotic changes at the Eocene–Oligocene transition in south-eastern Tibet. Natl. Sci. Rev. 2018, nwy062. [Google Scholar] [CrossRef]
- Wang, J.; Gu, W.; Xia., L.; Wan, J.; Gan, S. Geographical variation and provenance division of Alnus cremastogyne. J. Zhejiang For. Coll. 2005, 22, 502–506. (In Chinese) [Google Scholar] [CrossRef]
- Tijerino, A.; Korpelainen, H. Molecular characterization of Nicaraguan Pinus tecunumanii Schw. ex Eguiluz et Perry populations for in situ conservation. Trees 2014, 28, 1249–1253. [Google Scholar] [CrossRef]
- Gibson, J.P.; Rice, S.A.; Stucke, C.M. Comparison of population genetic diversity between a rare, narrowly distributed species and a common, widespread species of Alnus (Betulaceae). Am. J. Bot. 2008, 95, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.A.; Graves, W.R. Systematics of Alnus maritima (seaside alder) resolved by ISSR polymorphisms and morphological characters. J. Am. Soc. Hortic. Sci. 2004, 129, 231–236. [Google Scholar] [CrossRef]

| ID | Location | Sample Size | Longitude (E) | Latitude (N) | Elevation (m) |
|---|---|---|---|---|---|
| AA | Meigu | 10 | 103.08 | 28.30 | 1609–2352 |
| AC | Ebian | 13 | 103.25 | 29.28 | 828–1180 |
| B | Luding | 14 | 102.13 | 29.77 | 1303–2343 |
| F | Mianning | 16 | 102.10 | 29.18 | 1884–1908 |
| I | Pingwu | 11 | 104.50 | 32.30 | 845–1440 |
| J | Qingchuan | 12 | 105.14 | 32.41 | 552–1496 |
| K | Jiange | 13 | 105.45 | 32.16 | 569–808 |
| N | Bazhong | 24 | 106.63 | 31.68 | 368–591 |
| Q | Xuanhan | 10 | 107.62 | 31.55 | 482–998 |
| R | Yanting | 7 | 105.30 | 30.89 | 380–530 |
| S | Jintang | 12 | 103.80 | 30.74 | 740–960 |
| T | Qionglai | 9 | 103.16 | 30.32 | 746–1118 |
| U | Muchuan | 16 | 103.71 | 29.15 | 500–530 |
| V | Emeishan | 8 | 103.33 | 29.59 | 650–1273 |
| Locus | Repeat Motif | Primer Sequence (5’-3’) | Size Range | Tm (°C) | Na | Ne | Ho | Hs | H’t | G’st(Nei) | Nm | Primer Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acg3 | (CT)3CC(CT)2CC (CT)13AT(CT)5 | F: CTCCTTAGCTGGCACGGAC R: CCCTTCTTCATAAAACCCTCAA | 210–262 | 55 | 18 | 4.977 | 0.960 | 0.822 | 0.853 | 0.037 | 6.507 | L3.1 [9] |
| Acg7 | (AG)11 | F: CTGGGTTCAAACTTCCTTTGCT R: ACCCTAGGCCATGTTTGGTT | 265–293 | 56 | 13 | 3.770 | 0.949 | 0.761 | 0.779 | 0.024 | 10.167 | Alma7 [10] |
| Acg8 | (AG)12 | F: TGGCGACTATTAGAAGGACGA R: TGTGGGATAGCAAGTGTTGGA | 177–217 | 56 | 16 | 4.054 | 0.799 | 0.788 | 0.805 | 0.020 | 12.25 | Alma12 [10] |
| Acg11 | (GT)12(GA)10 | F: TGGGCTAGCATTAAGCACCA R: AAGGCCTCCCTTCCAACTTT | 248–256 | 58 | 5 | 2.244 | 0.891 | 0.580 | 0.577 | −0.006 | 1000 | Alma20 [10] |
| Acg16 | (AG)14 | F: GCAGACCAGAGTCTGTTATTCA R: AGACAATTTCGTGACTGGGTAT | 156–168 | 60 | 7 | 4.652 | 0.844 | 0.818 | 0.834 | 0.019 | 12.908 | CAC-A105 [18] |
| Acg19 | (TC)15 | F: CAGTCTATCTGCTACAAGCGTGGT R:GACGTTTTCAACGACCAAAAACAC | 130–138 | 58 | 5 | 2.674 | 0.902 | 0.652 | 0.669 | 0.025 | 9.75 | CAC-B113 [18] |
| Acg20 | (GA)12 | F: AAGCAAAATCCCAAGGTATCCAGT R: GGGGTTCCAACCAATTTATTCTTC | 142–196 | 58 | 25 | 2.464 | 0.660 | 0.621 | 0.631 | 0.016 | 15.375 | CAC-C118 [18] |
| Acg21 | (TC)12 | F: GATGGTAATGTGACGTGAGCAAAA R: CCTATTCTCATCGTTTAAAGCCCC | 247–287 | 58 | 20 | 6.084 | 0.987 | 0.859 | 0.870 | 0.014 | 17.607 | Ag01 [19] |
| Acg22 | (TC)11 | F: AACTTGTCTTATTGTGCACTTGCG R: ACATTTACGGCTAAACAGCATTCC | 184–210 | 58 | 8 | 3.092 | 0.956 | 0.701 | 0.713 | 0.017 | 14.456 | Ag05 [19] |
| Acg23 | (TG)12 | F: CAAGCGAAATAGATTCGTGGTCTT R: CTTCCATTTGGAGCCTTAAAACAC | 248–280 | 58 | 14 | 3.312 | 0.478 | 0.737 | 0.754 | 0.016 | 15.375 | Ag09 [19] |
| Ace1 | (TC)11n(TC)8ta (TC)6 | F: GCGCGCTCTCACTTTCTCTA R: CCACTTCTCCTCCTCGTCAC | 220–248 | 60 | 13 | 1.689 | 0.555 | 0.430 | 0.436 | 0.008 | 31.00 | FQ338662 [20] |
| Ace3 | (CT)12(CA)6 | F: TTCCTCAACAAAAACCCACC R: CCACGTCTCCCGAACACTAT | 210–240 | 60 | 11 | 3.786 | 0.935 | 0.760 | 0.792 | 0.039 | 6.16 | FQ335170 [20] |
| Ace27 | (CT)13(CA)7 | F: CCTTCTCACTTCACCCTCCA R: GAGGATGTGTGGCATTGTTG | 155–185 | 58 | 15 | 6.536 | 0.990 | 0.872 | 0.883 | 0.013 | 18.981 | FQ351578 [20] |
| Ace29 | (TC)26 | F: TGAATTTCTCCGGGACTTTG R: TCTCGGGAAATATCGACTTCA | 219–285 | 58 | 27 | 7.318 | 0.951 | 0.918 | 0.934 | 0.018 | 13.639 | FQ344263 [20] |
| Ace35 | (TC)20 | F: CGTGTGTGCCTTTGACCTTA R: ACTATTCAAGGAGGACGCGA | 194–200 | 58 | 4 | 1.682 | 0.681 | 0.431 | 0.459 | 0.060 | 3.917 | FQ334282 [20] |
| Ace37 | (GA)19 | F: CGGCAAGAACAACGAAGAAT R: AGCTGGAAGCTGATGACGAT | 119–137 | 58 | 10 | 1.402 | 0.376 | 0.307 | 0.315 | 0.005 | 49.75 | FQ351410 [20] |
| Act1 | (AG)5 | F: GGCCTGGATGTTGAGATAGC R: ATCAACTGACAACAGGCAACC | 122–134 | 60 | 4 | 1.846 | 0.654 | 0.483 | 0.541 | 0.106 | 2.108 | [20] |
| Act7 | (GT)5 | F: CAGCGACTAACACTGCATGA R: TATTGCTCAAAGGGAGCAGC | 239–245 | 59 | 4 | 1.640 | 0.476 | 0.414 | 0.406 | −0.020 | 1000 | [20] |
| Act9 | (TA)5 | F: TTTGTGGTTGGTTGAAGGTC R: AAAGGGTGAAGACGTGGATG | 204–212 | 59 | 5 | 1.529 | 0.605 | 0.366 | 0.362 | −0.011 | 1000 | [20] |
| Act12 | (AT)7 | F: CACGTACGCACGCATGTA R: TGAGTTCATTATCTTCGTGATTGA | 124–150 | 59 | 14 | 3.846 | 0.479 | 0.822 | 0.871 | 0.055 | 4.295 | [20] |
| Act15 | (TC)5 | F: TCGAAGTACACAATTTGCCA R: TTCTTGTCCTCTTTGGATTCG | 245–251 | 59 | 4 | 1.980 | 0.812 | 0.521 | 0.504 | −0.035 | 1000 | [20] |
| Act23 | (TA)5 | F: GGAGCTAGTCGAGGCATTTC R: TTTGGGTTCAGAGCTTCACTC | 219–227 | 59 | 5 | 2.045 | 0.862 | 0.536 | 0.536 | −0.002 | 1000 | [20] |
| Act27 | (CT)7 | F: GCTTCTCTGCTGTTTGGTCC R: TGTGCGAAACTCGCAAATTA | 217–227 | 60 | 6 | 1.649 | 0.288 | 0.423 | 0.452 | 0.064 | 3.656 | [20] |
| Act29 | (AT)6 | F: ATTCCCGACTGTCTCAAAGC R: CTCTTTGATCCTTTGTTCTTTGG | 147–153 | 59 | 4 | 2.608 | 0.857 | 0.641 | 0.660 | 0.029 | 8.371 | [20] |
| Act30 | (TA)5 | F: GGTGGTTCGAACACTCCTCT R: CATTCACTTACCAATTGCCC | 103–131 | 59 | 14 | 3.162 | 0.535 | 0.730 | 0.745 | 0.016 | 15.375 | [20] |
| Mean | 10.84 | 3.202 | 0.739 | 0.640 | 0.655 | 0.021 | 11.114 |
| Population | Na | Ne | Ho | Hs | Ht | Gis |
|---|---|---|---|---|---|---|
| AA | 6.92 | 3.947 | 0.762 | 0.681 | 0.681 | 0.005 |
| AC | 5.68 | 3.301 | 0.724 | 0.623 | 0.623 | −0.036 |
| B | 6.28 | 3.615 | 0.711 | 0.643 | 0.643 | 0.020 |
| F | 7.80 | 3.962 | 0.794 | 0.682 | 0.682 | −0.038 |
| I | 5.36 | 2.905 | 0.735 | 0.608 | 0.608 | −0.055 |
| J | 5.04 | 3.139 | 0.758 | 0.615 | 0.615 | −0.083 |
| K | 5.68 | 3.266 | 0.709 | 0.605 | 0.605 | −0.029 |
| N | 6.72 | 3.450 | 0.712 | 0.606 | 0.606 | −0.037 |
| Q | 5.64 | 3.307 | 0.741 | 0.621 | 0.621 | −0.050 |
| R | 4.16 | 2.842 | 0.712 | 0.593 | 0.593 | −0.046 |
| S | 5.48 | 3.277 | 0.725 | 0.617 | 0.617 | −0.043 |
| T | 5.08 | 3.297 | 0.800 | 0.656 | 0.656 | −0.116 |
| U | 6.68 | 3.611 | 0.706 | 0.629 | 0.629 | −0.007 |
| V | 5.04 | 3.261 | 0.761 | 0.637 | 0.637 | −0.071 |
| Mean | 5.83 | 3.37 | 0.739 | 0.630 | 0.630 | −0.042 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.-Y.; Wang, Z.-L.; Huang, Z.; Chen, Z.; Yang, H.-B.; Kang, X.-Y. Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers. Forests 2019, 10, 278. https://doi.org/10.3390/f10030278
Guo H-Y, Wang Z-L, Huang Z, Chen Z, Yang H-B, Kang X-Y. Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers. Forests. 2019; 10(3):278. https://doi.org/10.3390/f10030278
Chicago/Turabian StyleGuo, Hong-Ying, Ze-Liang Wang, Zhen Huang, Zhi Chen, Han-Bo Yang, and Xiang-Yang Kang. 2019. "Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers" Forests 10, no. 3: 278. https://doi.org/10.3390/f10030278





