A Distributed Indexing Method for Timeline Similarity Query
Abstract
:1. Introduction
- The triangle increment partition strategy (TIPS) is proposed to support the representation and partition of the interval dataset for indexing timeline datasets.
- The DTI-Tree based on TIPS is proposed to effectively and efficiently index timelines. DTI-Tree is a novel indexing method for interval-based discrete timelines similarity query based on Apache Spark.
- The evaluation function of timeline similarity has been presented and the timeline similarity query has been implemented by the supporting of the DTI-Tree.
- An open source timeline test dataset generator named TimelineGenerator is developed. It can generate various timeline test datasets for different conditions, and may provide some benchmark timeline datasets for the future research.
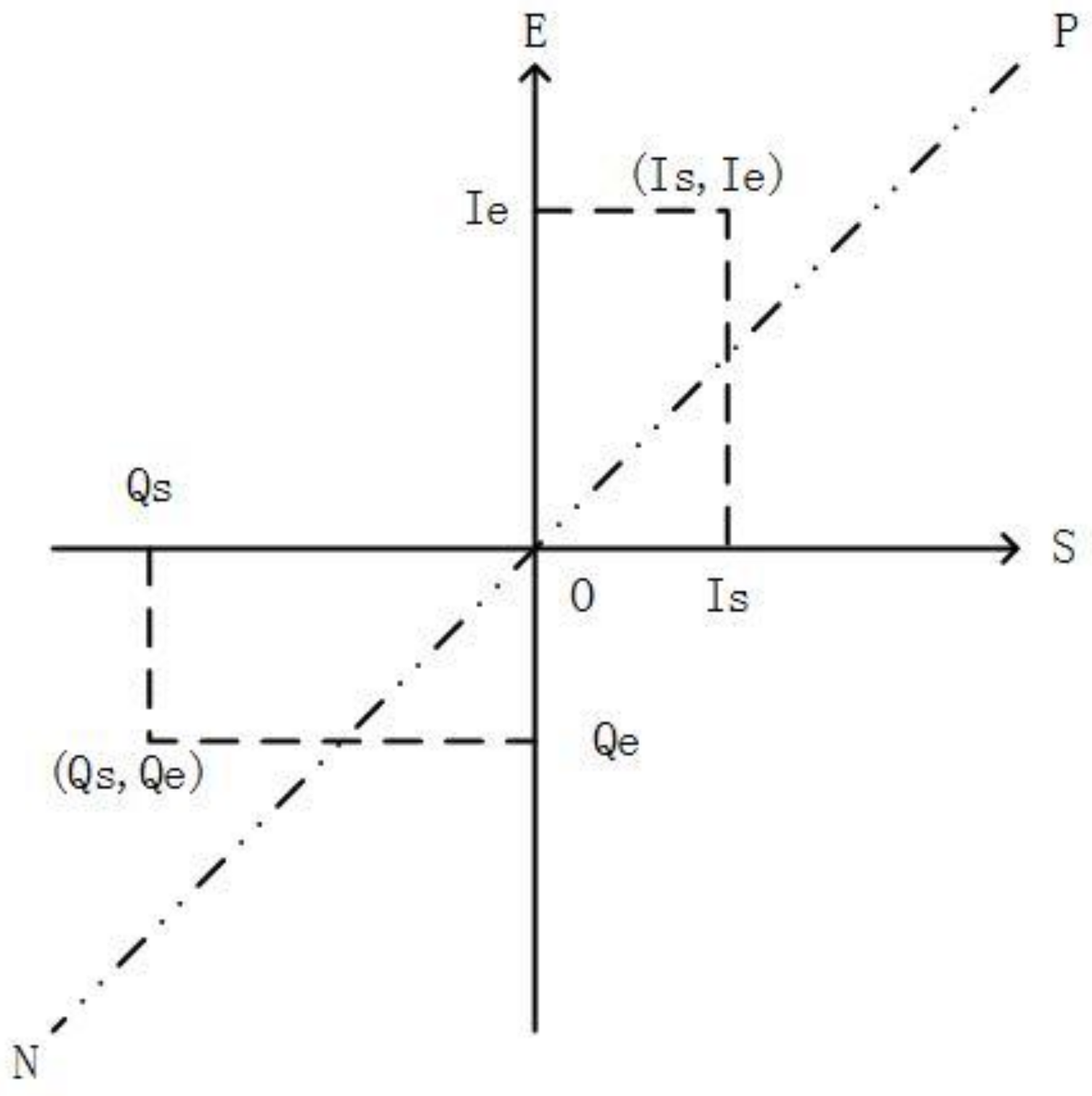
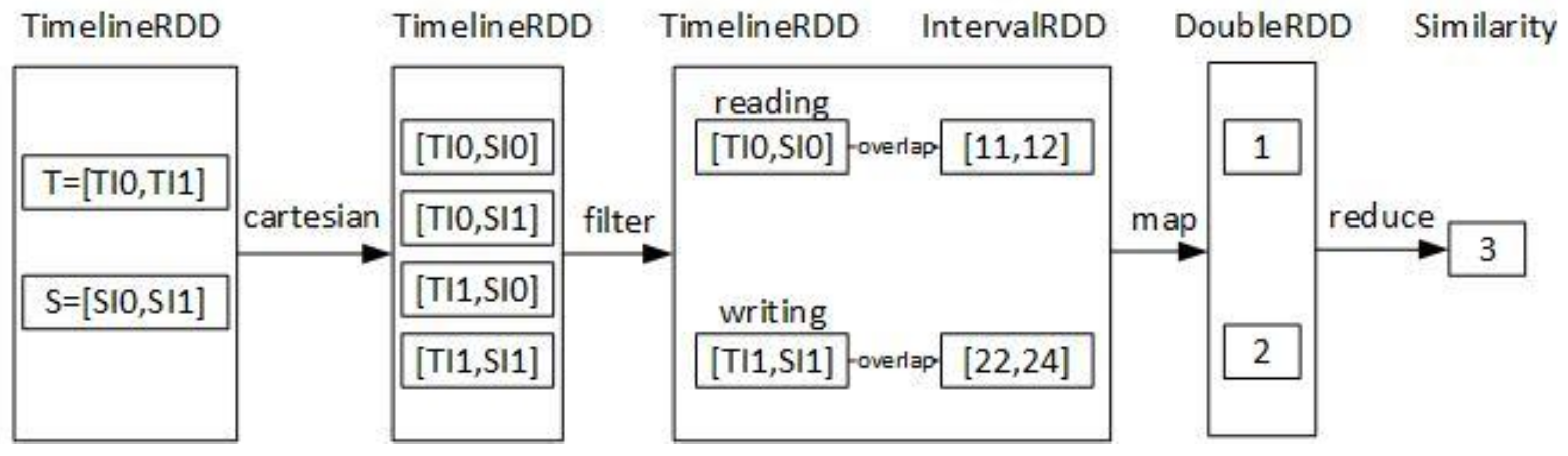
2. Timeline’s Representation and Similarity Function
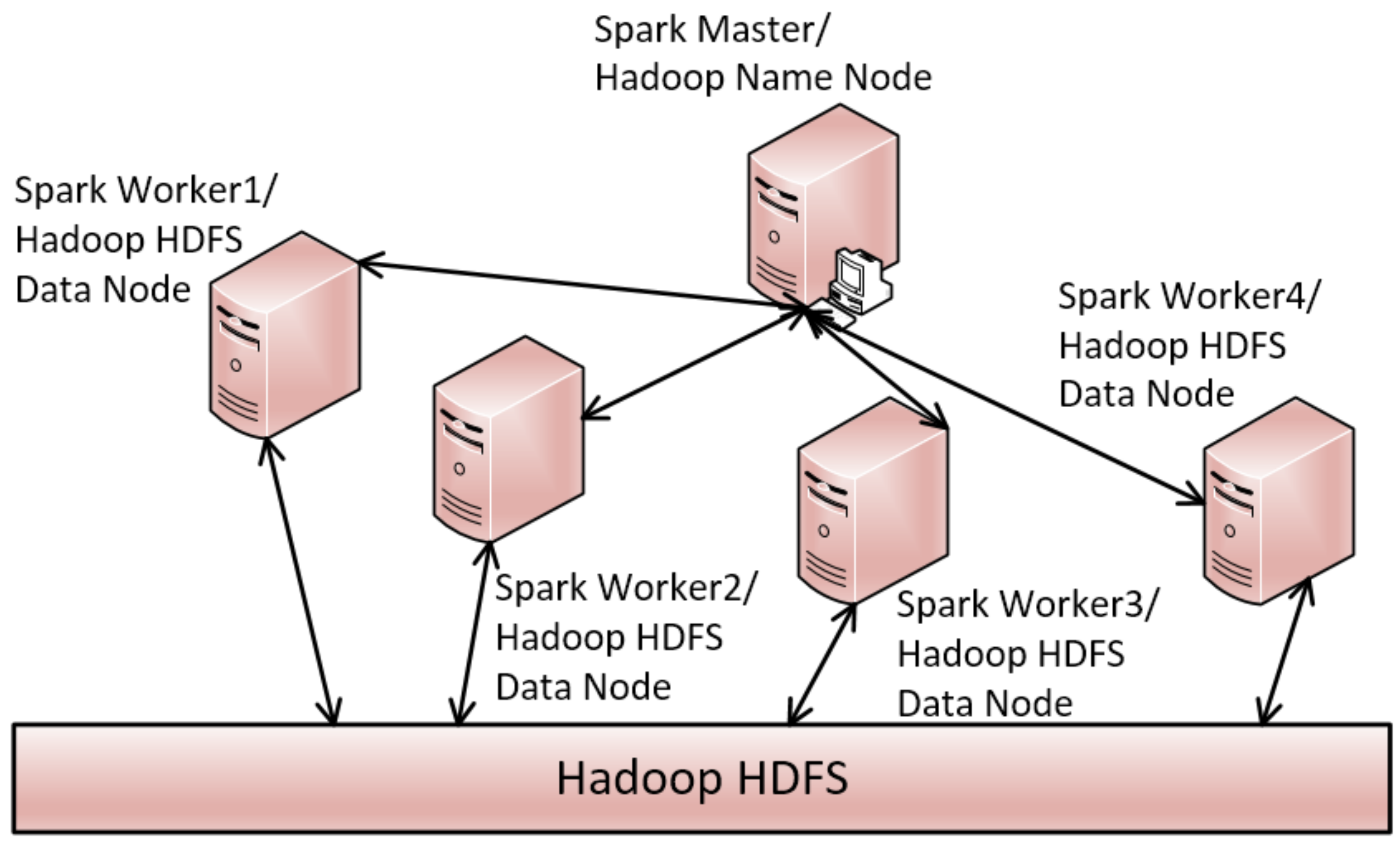
3. Distributed Triangle Increment Tree (DTI-Tree)
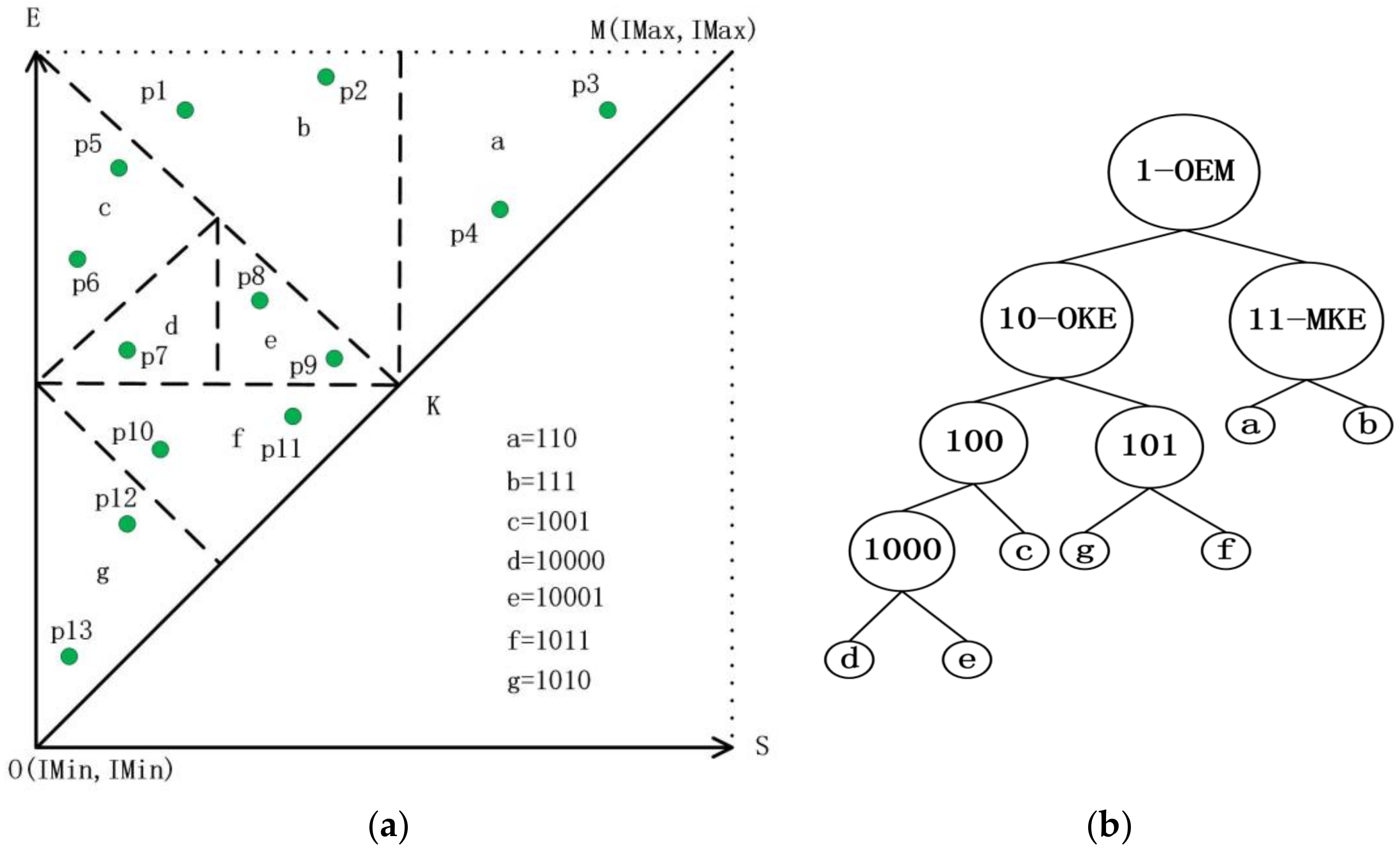
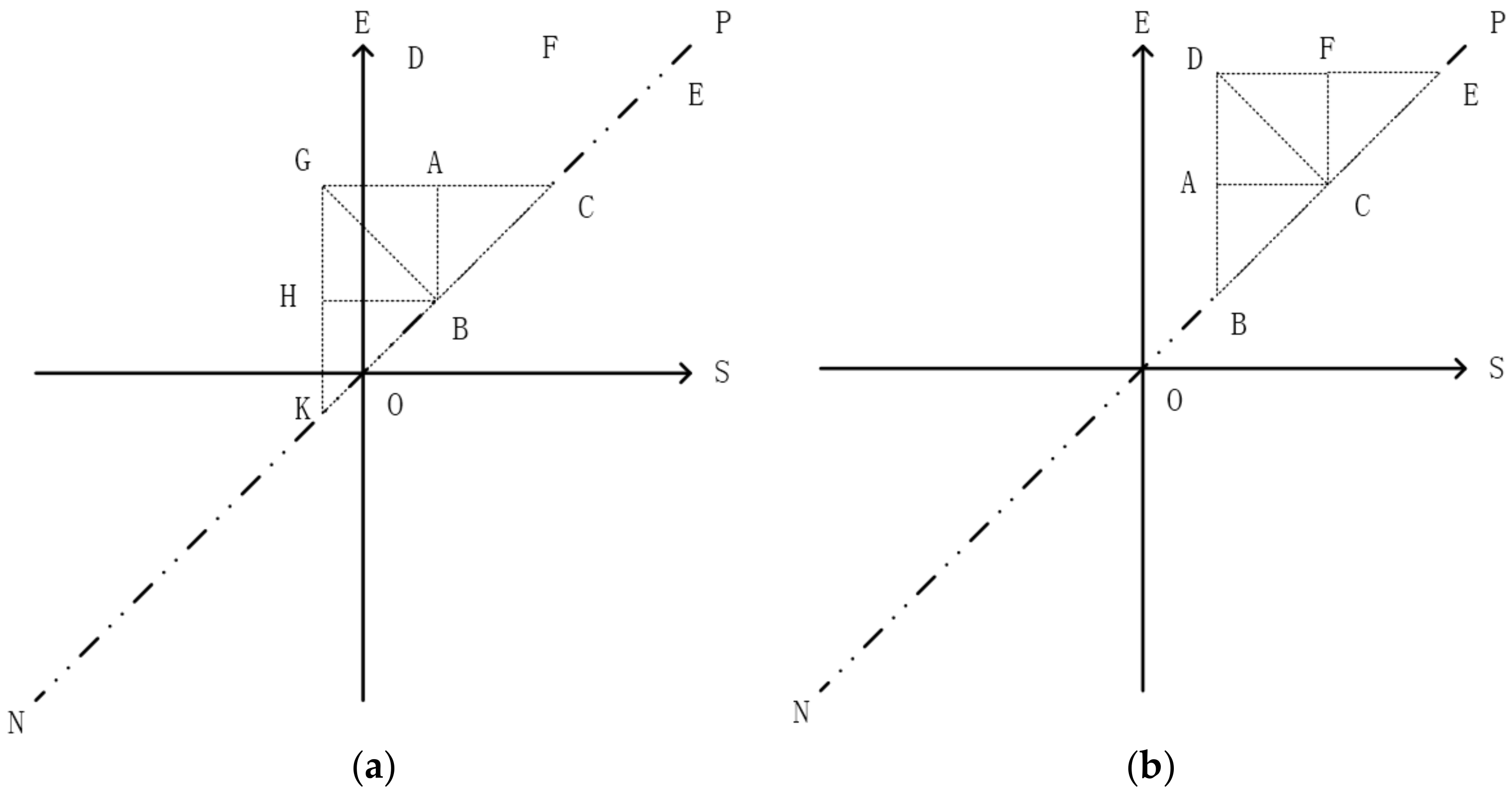
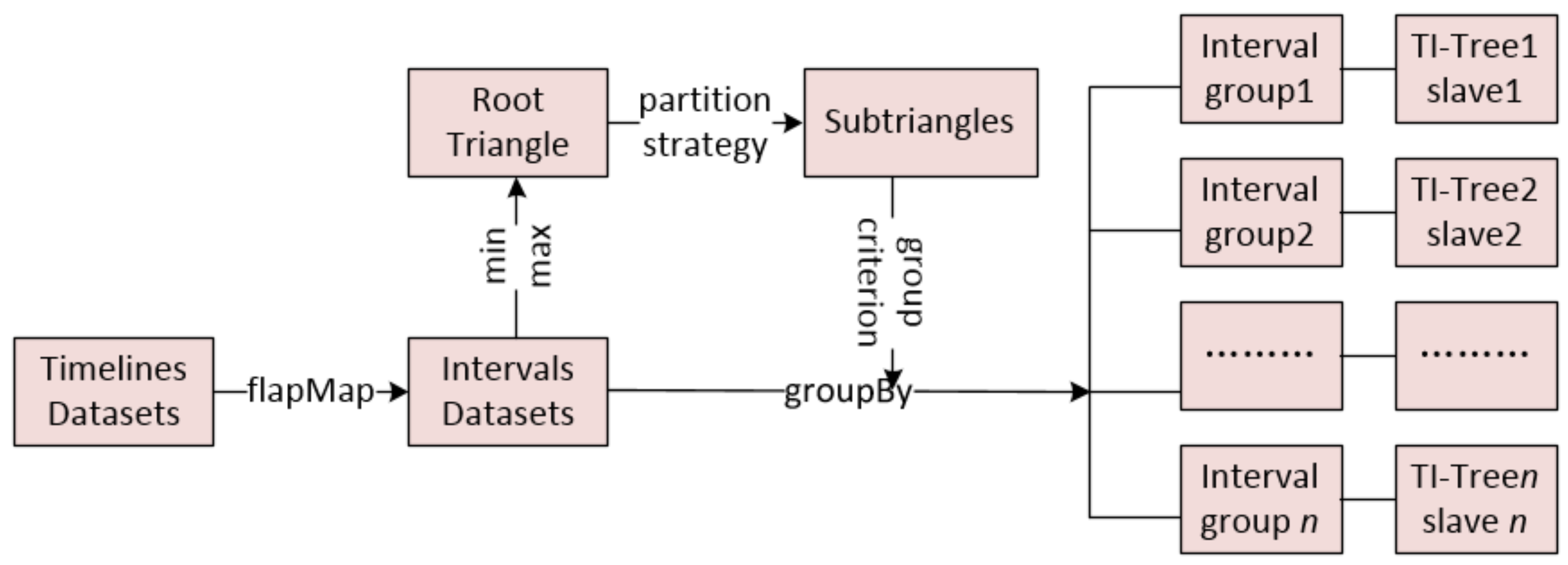
3.1. The Strategy of Partition
| Algorithm 1: Tips |
| Input: pi: an interval [Is, Ie]; root: root node of TI-Tree or T-Tree; Output: root: the new root node { 1. read the root node from disk; 2. if (root._triangle contains pi) return root; 3. if (pi is located at the left of line AB) executes left extension shown in Figure 8a; else executes right extension shown in Figure 8b; 4. while (root. _triangle does not contain pi) call Algorithm 1 recursively; 5. return root; } |
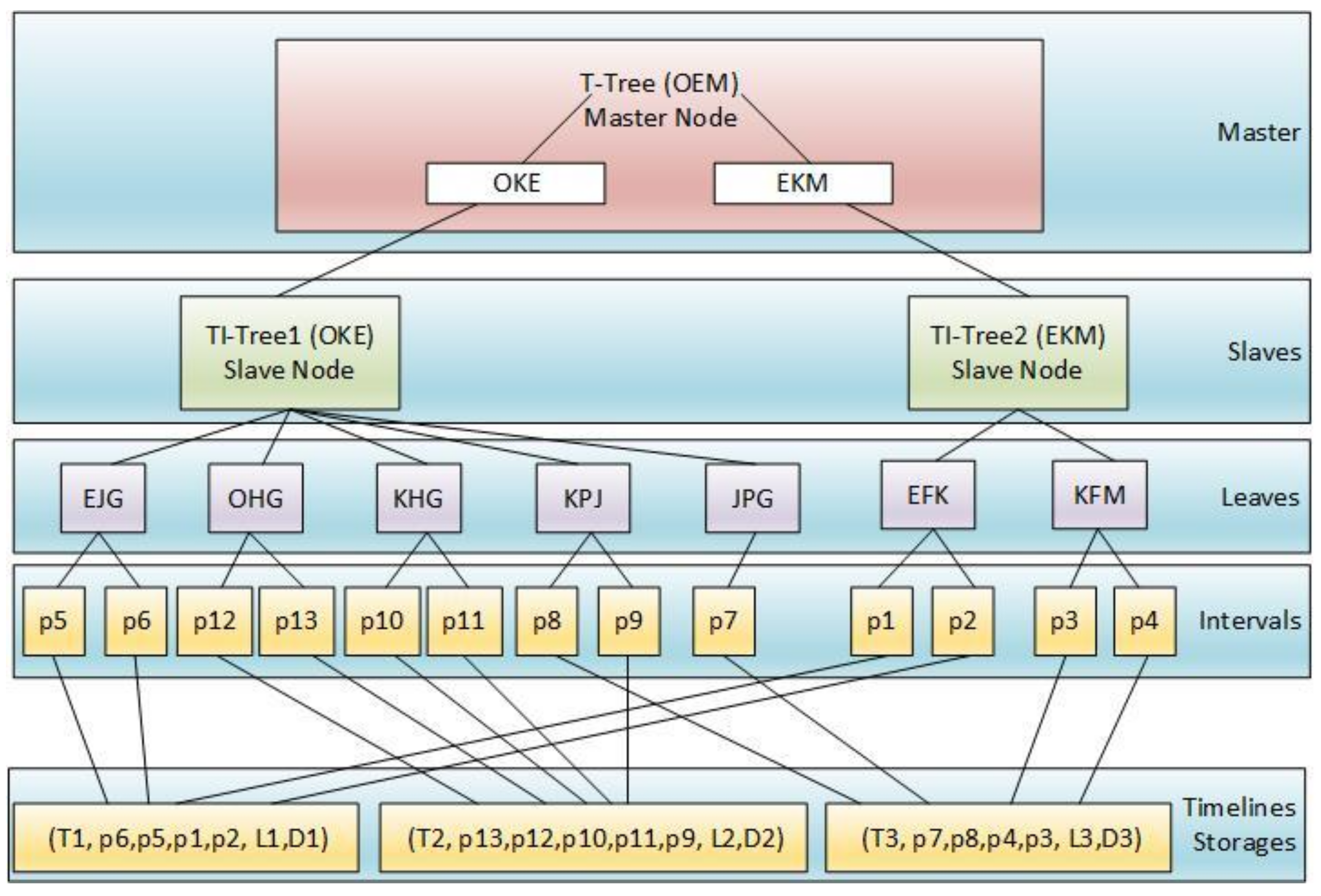
4. The Structure of DTI-Tree
4.1. The Construction of DTI-Tree
| Algorithm 2: TI-Tree construction |
| Input: IDS: an interval data set; Output: TI-Tree { 1. initial the root triangle variable root; 2. i = 0; 3. while (i < IDS.size()) if (root._triangle.constains(IDS [i])) call the TD-Tree’s insertion procedure; i++; else call Algorithm 1; 4. return TI-Tree. } |
| Algorithm 3: DTI-Tree construction shown in Figure 10 |
| Input: TD: a timeline data set; Output: DTI-Tree { 1. IDS= flapMap(TD); //convert timeline set into interval set. 2. initialize and recursively binary split the root triangle by TIPS until the number of the leaf node triangle is equal to the number of all the slaves SN in the cluster; 3. call the function groupBy to split IDS into SN groups according to the leaf nodes’ triangles generated in Step 2; 4. executes load balance evaluation and avoid adjustment after construction; 5. for each group, call Algorithm 2 to construct TI-Tree on the corresponding slave; } |
4.2. The Update of DTI-Tree
| Algorithm 4: DTI-Tree insertion |
| Input: T: a timeline; Output: DTI-Tree { 1. call the function intervalize listed in Table 3 to get a set of interval points IPS; 2. select the minimum triangle minTri which contains IPS; 3. get the slaves whose root triangle is intersected with minTri; 4. if (the number of slaves is equal 1) let the corresponding TI-Tree insert all the interval points of the timeline else divide the IPS into several parts; let the corresponding TI-Tree insert each part. } |
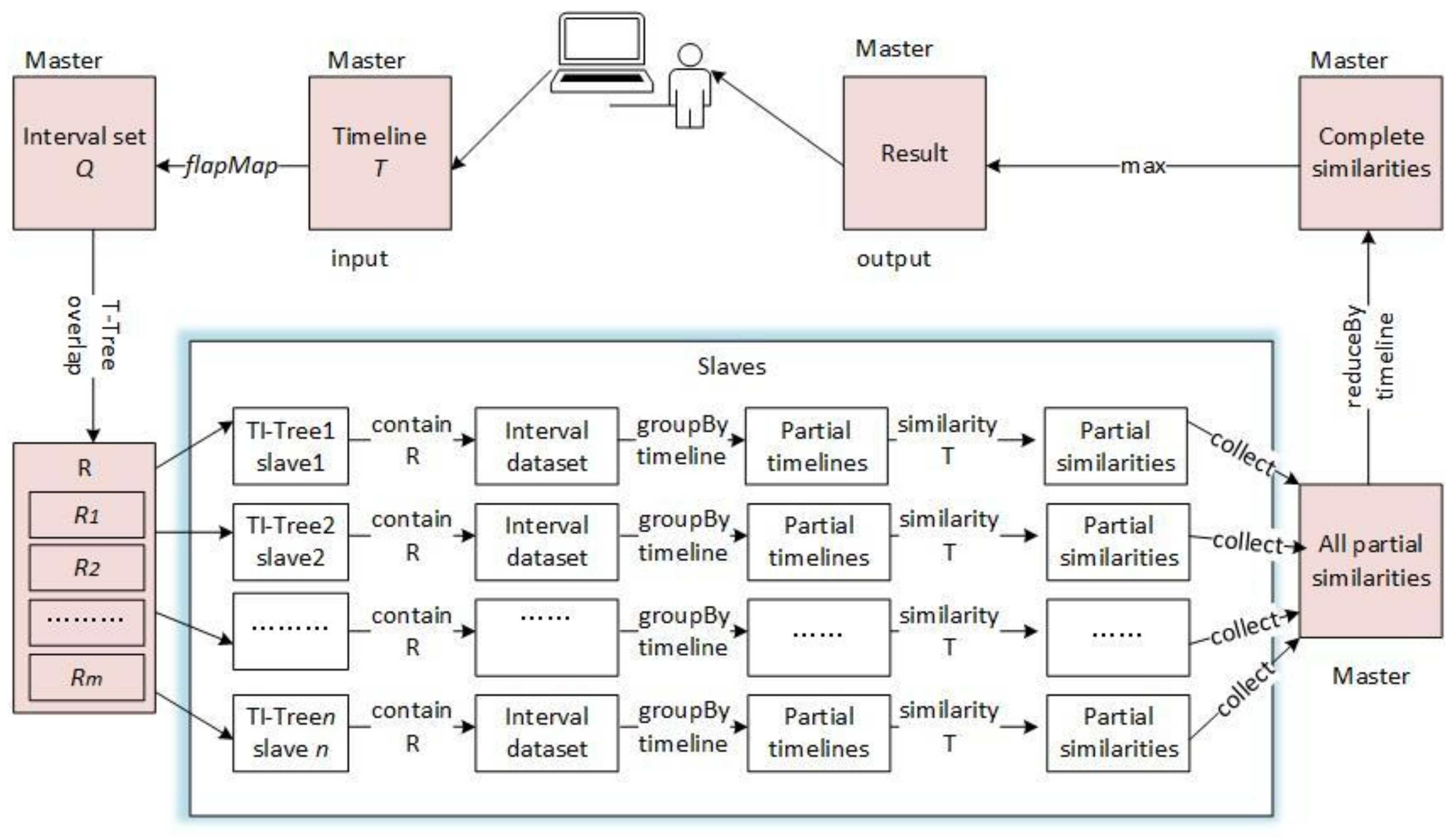
4.3. The Timeline Similarity Query of DTI-Tree
| Algorithm 5: DTI-Tree similarity query shown in Figure 11 |
| Input: T: a timeline; Output: the most similar timeline to T { 1. call the function intervalize to get a set of interval points Q = {Q0, Q1, …, Qk, …, Qm}; 2. calculate the overlap query region of Qk, noted as Rk; R = R0 R1, …, Rk, …, Rm; 3. find all the slave root triangles which overlaps R by the T-Tree in the master node, the result is {RTi}. RTi is the root triangle of the TI-Treei on the slave node Si. 4. find all the interval points Pi in the rectangle R by the TI-Treei on the slave Si separately; 5. execute Pi .groupBy(timeline ID) on the slave Si separately, and the result is some partial timelines PTi={PTij}; 6. execute similarity (T, PTij) and send the result that are some partial similarity values PSVi to the master; 7. combine all the partial similarity values {PSVij} on the master node, here i is a slave identifier and j is a timeline identifier; 8. return the result of {PSVij}. reduceBy(j). max (); } |
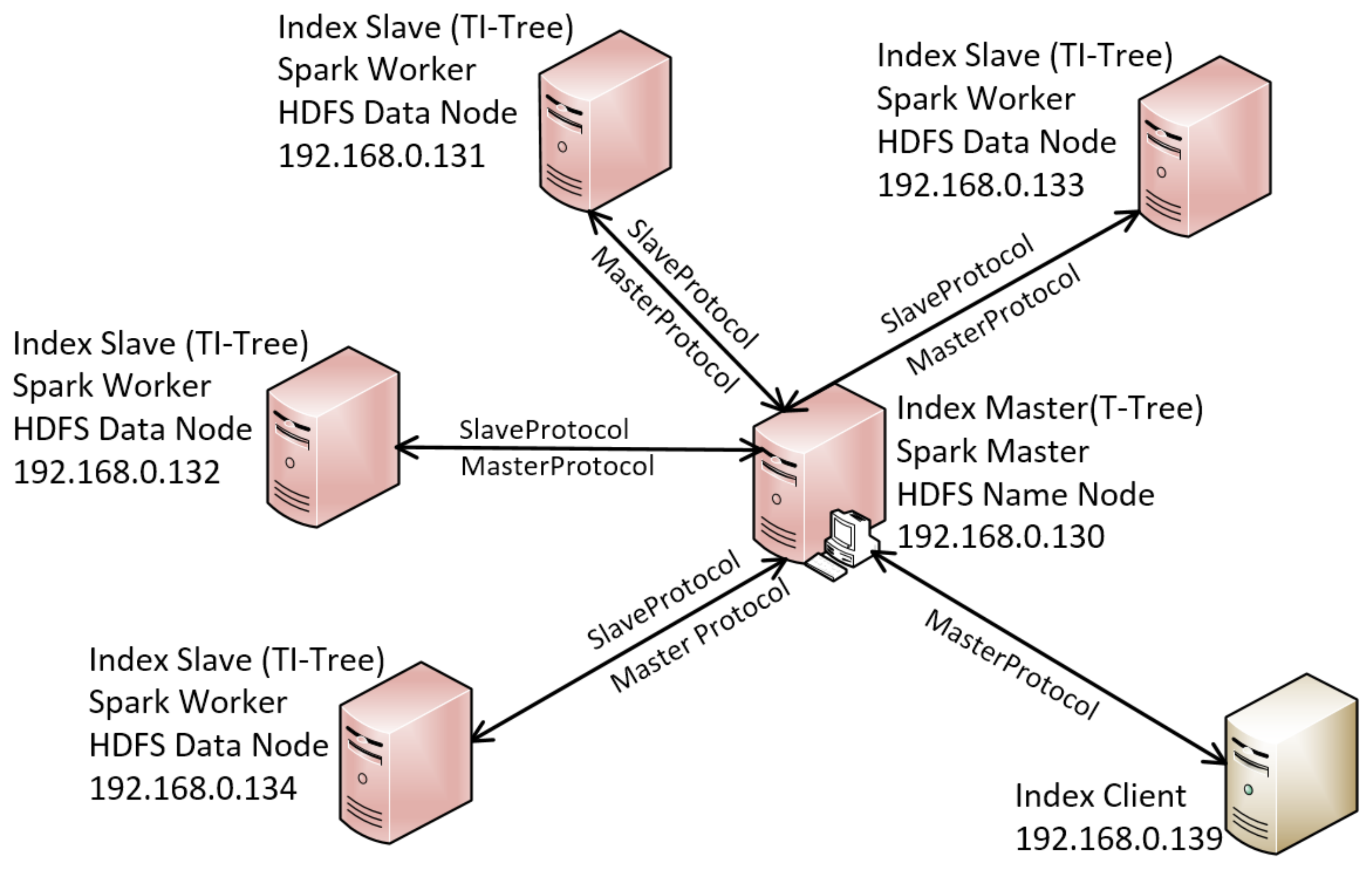
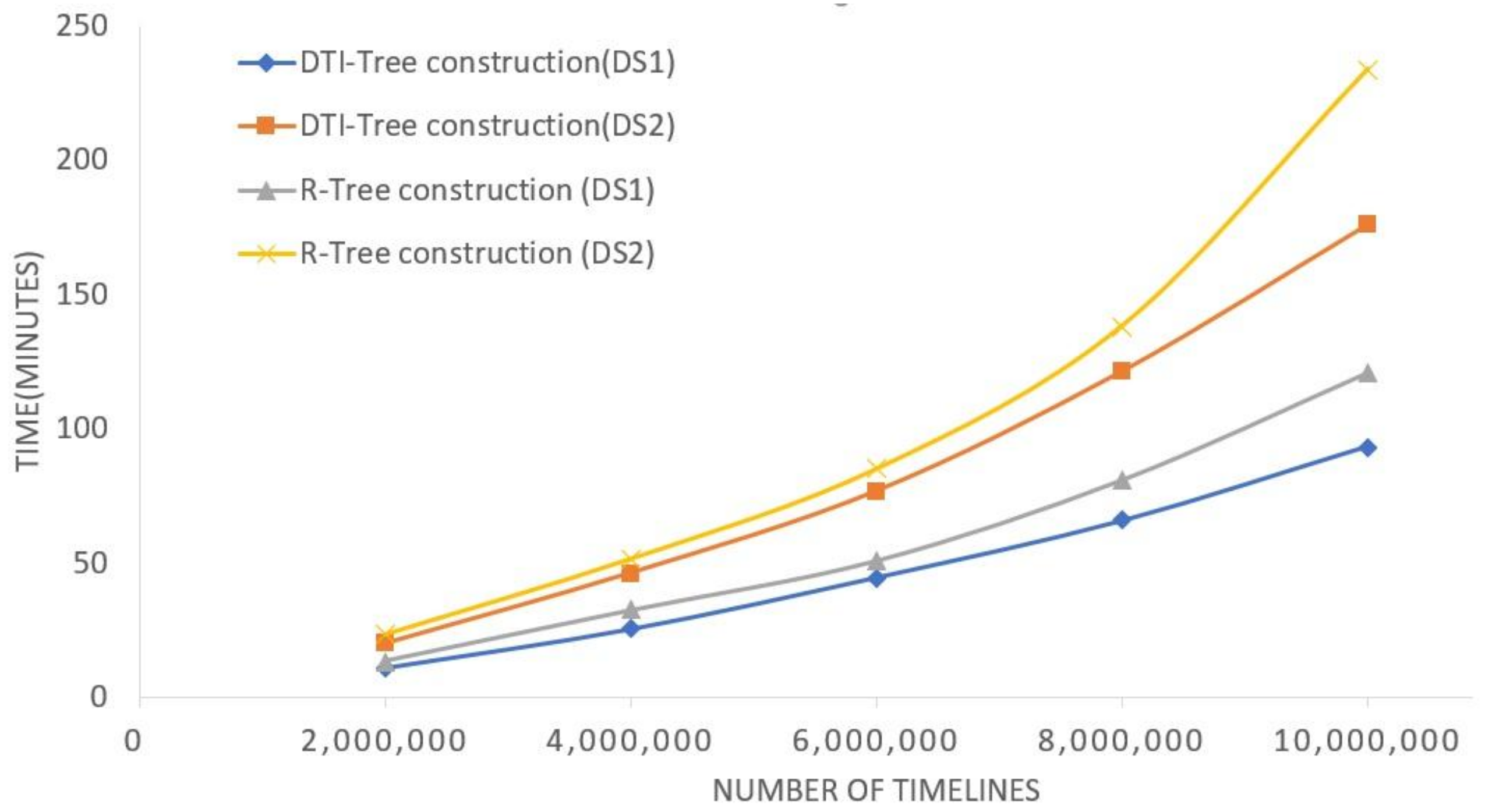
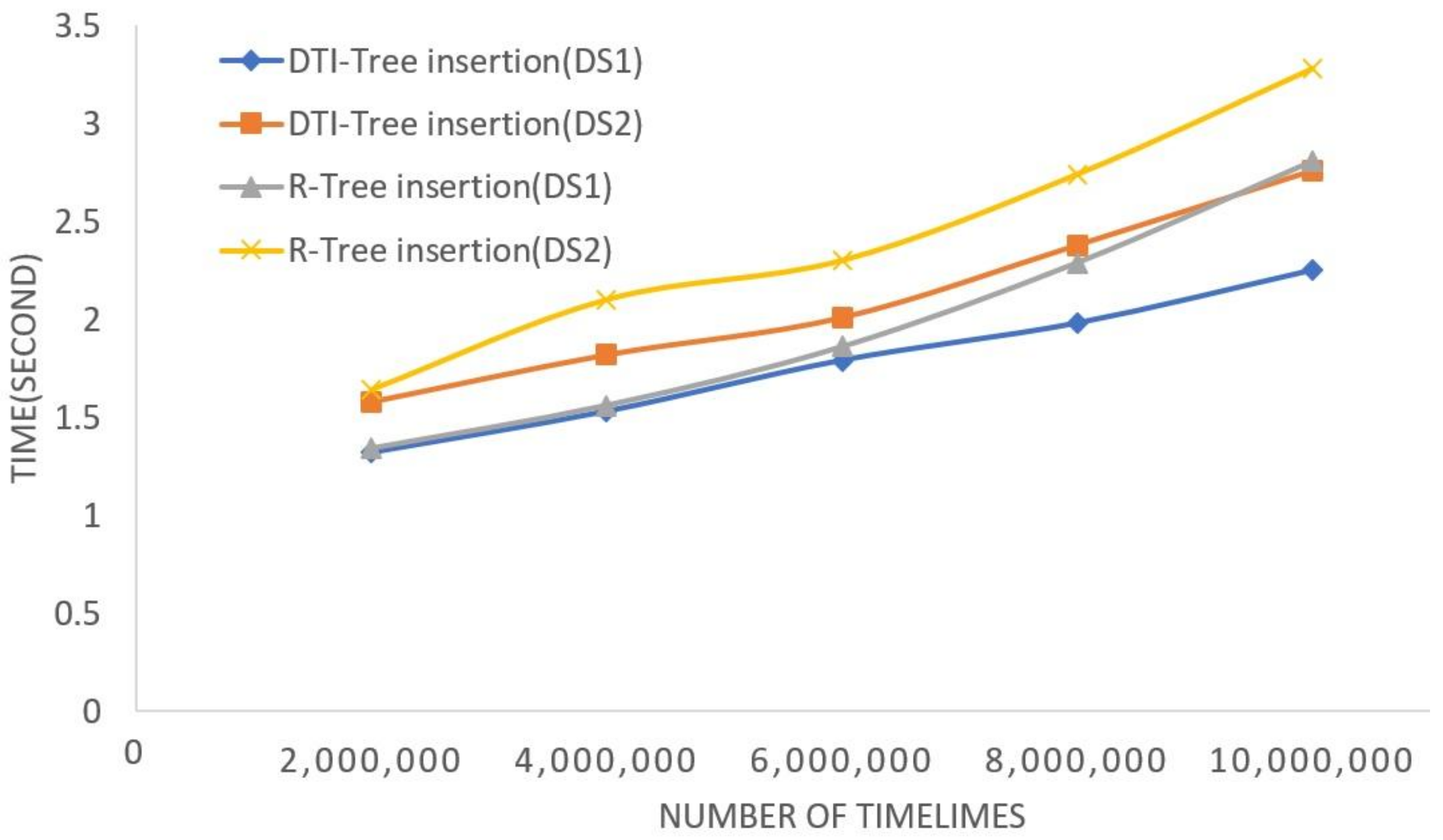
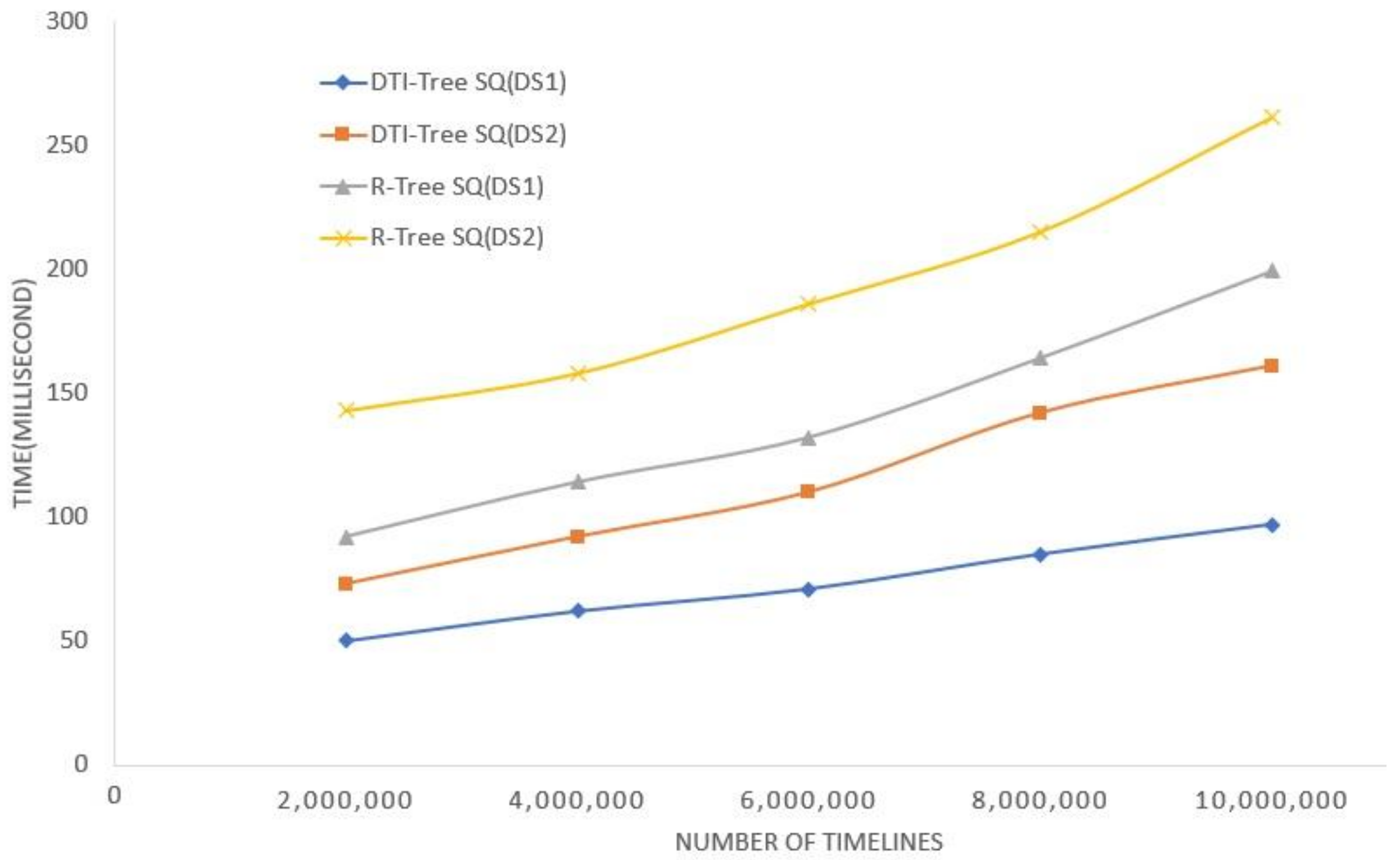
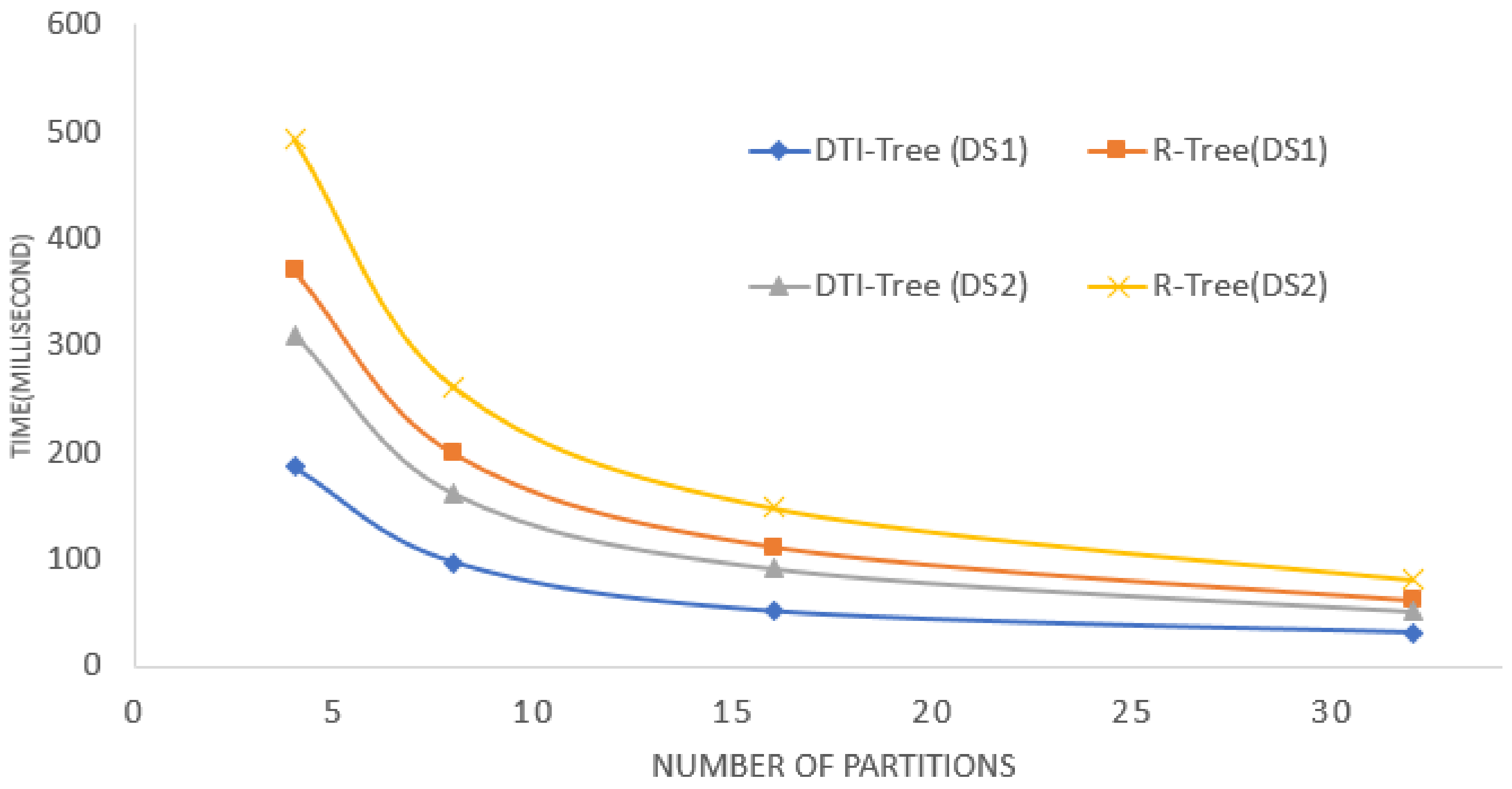
5. Experimental Evaluation
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brehmer, M.; Lee, B.; Bach, B.; Riche, N.H.; Munzner, T. Timelines revisited: A design space and considerations for expressive storytelling. IEEE Trans. Vis. Comput. Graph. 2017, 23, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Camerra, A.; Palpanas, T.; Shieh, J.; Keogh, E. Isax 2.0: Indexing and Mining One Billion Time Series. In Proceedings of the 2010 10th IEEE International Conference on Data Mining (ICDM), Sydney, NSW, Australia, 14–17 December 2010; Institute of Electrical and Electronics Engineers Inc.: Sydney, NSW, Australia, 2010; pp. 58–67. [Google Scholar]
- Zoumpatianos, K.; Idreos, S.; Palpanas, T. Ads: The adaptive data series index. VLDB J. 2016, 25, 843–866. [Google Scholar] [CrossRef]
- Yagoubi, D.E.; Akbarinia, R.; Masseglia, F.; Palpanas, T. Dpisax: Massively distributed partitioned isax. In Proceedings of the 2017 17th IEEE International Conference on Data Mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; IEEE Computer Society Press: New Orleans, LA, USA, 2017; pp. 1135–1140. [Google Scholar]
- Kondylakis, H.; Dayan, N.; Zoumpatianos, K.; Palpanas, T. Coconut: A scalable bottom-up approach for building data series indexes. Proc. VLDB Endow. 2018, 11, 677–690. [Google Scholar]
- Kriegel, H.P.; Potke, M.; Seidl, T. Interval sequences: An object-relational approach to manage spatial data. In Proceedings of the 2001 7th International Symposium Advances in Spatial and Temporal Databases (SSTD), Redondo Beach, CA, USA, 12–15 July 2001; Jensen, C.S., Schneider, M., Seeger, B., Tsotras, V., Eds.; Springer: Berlin, Germany, 2001; Volume 2121, pp. 481–501. [Google Scholar]
- Kriegel, H.-P.; Pötke, M.; Seidl, T.; Jensen, C.; Schneider, M.; Seeger, B.; Tsotras, V. Object-relational indexing for general interval relationships. In Advances in Spatial and Temporal Databases; Springer: Berlin/Heidelberg, Germany, 2001; Volume 2121, pp. 522–542. [Google Scholar]
- Stantic, B.; Topor, R.; Terry, J.; Sattar, A. Advanced indexing technique for temporal data. Comput. Sci. Inf. Syst. 2010, 7, 679–703. [Google Scholar] [CrossRef]
- He, Z.; Kraak, M.-J.; Huisman, O.; Ma, X.; Xiao, J. Parallel indexing technique for spatio-temporal data. ISPRS J. Photogramm. Remote Sens. 2013, 78, 116–128. [Google Scholar] [CrossRef]
- Ma, X.; Fox, P. Recent progress on geologic time ontologies and considerations for future works. Earth Sci. Inform. 2013, 6, 31–46. [Google Scholar] [CrossRef]
- Ma, X.; Carranza, E.J.M.; Wu, C.; van der Meer, F.D.; Liu, G. A skos-based multilingual thesaurus of geological time scale for interoperability of online geological maps. Comput. Geosci. 2011, 37, 1602–1615. [Google Scholar] [CrossRef]
- He, Z.; Wu, C.; Liu, G.; Zheng, Z.; Tian, Y. Decomposition tree: A spatio-temporal indexing method for movement big data. Clust. Comput. 2015, 18, 1481–1492. [Google Scholar] [CrossRef]
- Xie, X.; Mei, B.; Chen, J.; Du, X.; Jensen, C.S. Elite: An elastic infrastructure for big spatiotemporal trajectories. VLDB J. 2016, 25, 473–493. [Google Scholar] [CrossRef]
- Elmasri, R.; Wuu, G.; Kim, Y. The Time Index: An Access Structure for Temporal Data. In Proceedings of the 16th International Conference on Very Large Databases, Brisbane, Australia, 13–16 August 1990; pp. 1–12. [Google Scholar]
- Curtis, P.K.; Michael, S. Segment Indexes: Dynamic Indexing Techniques for Multi-Dimensional Interval Data. SIGMOD Rec. 1991, 20, 138–147. [Google Scholar]
- Chaabouni, M.; Chung, S.M. The point-range tree: A Data Structure for Indexing Intervals. In Proceedings of the 1993 ACM Conference on Computer Science, Washington, DC, USA, 25–28 May 1993; ACM: Indianapolis, IN, USA, 1993; pp. 453–460. [Google Scholar]
- Ang, C.H.; Tan, K.P. The interval B-tree. Inf. Process. Lett. 1995, 53, 85–89. [Google Scholar] [CrossRef]
- Tsotras, V.J.; Gopinath, B.; Hart, G.W. Efficient management of time-evolving databases. IEEE Trans. Knowl. Data Eng. 1995, 7, 591–608. [Google Scholar] [CrossRef]
- Lanka, S.; Mays, E. Fully Persistent B+-Trees. SIGMOD Rec. 1991, 20, 426–435. [Google Scholar] [CrossRef]
- Brodal, G.S.; Brodal, L.; Tsakalidis, K.; Sioutas, S.; Tsichlas, K. Fully Persistent B-Trees. In Proceedings of the 23rd annual ACM-SIAM Symposium on Discrete algorithms, Kyoto, Japan, 17–19 January 2012; Society for Industrial and Applied Mathematics: Kyoto, Japan, 2012; pp. 602–614. [Google Scholar]
- Kriegel, H.-P.; Pötke, M.; Seidl, T. Managing intervals efficiently in object-relational databases. In Proceedings of the 26th International Conference on Very Large Data Bases, San Francisco, CA, USA, 10–14 September 2000; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2000; pp. 407–418. [Google Scholar]
- Enderle, J.; Schneider, N.; Seidl, T. Efficiently processing queries on interval-and-value tuples in relational databases. In Proceedings of the 31st International Conference on Very Large Data Bases, Trondheim, Norway, 30 August–2 September 2005; VLDB Endowment: Trondheim, Norway, 2005; pp. 385–396. [Google Scholar]
- Wang, M.; Xiao, M. SHB+-tree: A segmentation hybrid index structure for temporal data. In Proceedings of the 2015 15th International Conference on Algorithms and Architectures for Parallel (ICA3PP), Zhangjiajie, China, 18–20 November 2015; Wang, G., Zomaya, A., Martinez Perez, G., Li, K., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 282–296. [Google Scholar]
- Bayer, R.; McCreight, E.M. Organization and maintenance of large ordered indexes. Acta Inform. 1972, 1, 173–189. [Google Scholar] [CrossRef]
- Douglas, C. Ubiquitous B-tree. ACM Comput. Surv. (CSUR) 1979, 11, 121–137. [Google Scholar]
- Guttman, A. R-trees: A dynamic index structure for spatial searching. SIGMOD Rec. 1984, 14, 47–57. [Google Scholar] [CrossRef]
- Kim, J.; Nam, B. Parallel multi-dimensional range query processing with R-trees on GPU. J. Parallel Distrib. Comput. 2013, 73, 1195–1207. [Google Scholar] [CrossRef]
- Eldawy, A.; Mokbel, M.F. Spatialhadoop: A mapreduce framework for spatial data. In Proceedings of the 2015 31st IEEE International Conference on Data Engineering, Seoul, Korea, 13–17 April 2015; IEEE Computer Society: Seoul, Korea, 2015; pp. 1352–1363. [Google Scholar]
- Dean, J.; Ghemawat, S. Mapreduce: Simplified data processing on large clusters. Commun. ACM 2008, 51, 107–113. [Google Scholar] [CrossRef]
- Aji, A.; Wang, F.; Vo, H.; Lee, H.; Liu, Q.; Zhang, X.; Saltz, J. Hadoop gis: A high performance spatial data warehousing system over mapreduce. Proc. VLDB Endow. 2013, 6, 1009–1020. [Google Scholar] [CrossRef]
- Alarabi, L.; Mokbel, M.F.; Musleh, M. St-hadoop: A mapreduce framework for spatio-temporal data. In Proceedings of the 2017 15th International Symposium Advances in Spatial and Temporal Databases (SSTD), Arlington, VA, USA, 21–23 August 2017; Gertz, M., Renz, M., Zhou, X., Hoel, E., Ku, W.-S., Voisard, A., Zhang, C., Chen, H., Tang, L., Huang, Y., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 84–104. [Google Scholar]
- Yu, J.; Wu, J.X.; Sarwat, M. Geospark: A Cluster Computing Framework for Processing Large-Scale Spatial Data; Assoc Computing Machinery: New York, NY, USA, 2015. [Google Scholar]
- Baig, F.; Mehrotra, M.; Vo, H.; Wang, F.; Saltz, J.; Kurc, T. Sparkgis: Efficient comparison and evaluation of algorithm results in tissue image analysis studies. In Proceedings of the Biomedical Data Management and Graph Online Querying: VLDB 2015 Workshops, Big-O(Q) and DMAH, Waikoloa, HI, USA, 31 August–4 September 2015; Wang, F., Luo, G., Weng, C., Khan, A., Mitra, P., Yu, C., Eds.; Revised Selected Papers. Springer International Publishing: Cham, Switzerland, 2016; pp. 134–146. [Google Scholar]
- Wang, H.; Belhassena, A. Parallel trajectory search based on distributed index. Inf. Sci. 2017, 388–389, 62–83. [Google Scholar] [CrossRef]
- Jalili, V.; Matteucci, M.; Masseroli, M.; Ceri, S. Indexing next-generation sequencing data. Inf. Sci. 2017, 384, 90–109. [Google Scholar] [CrossRef]
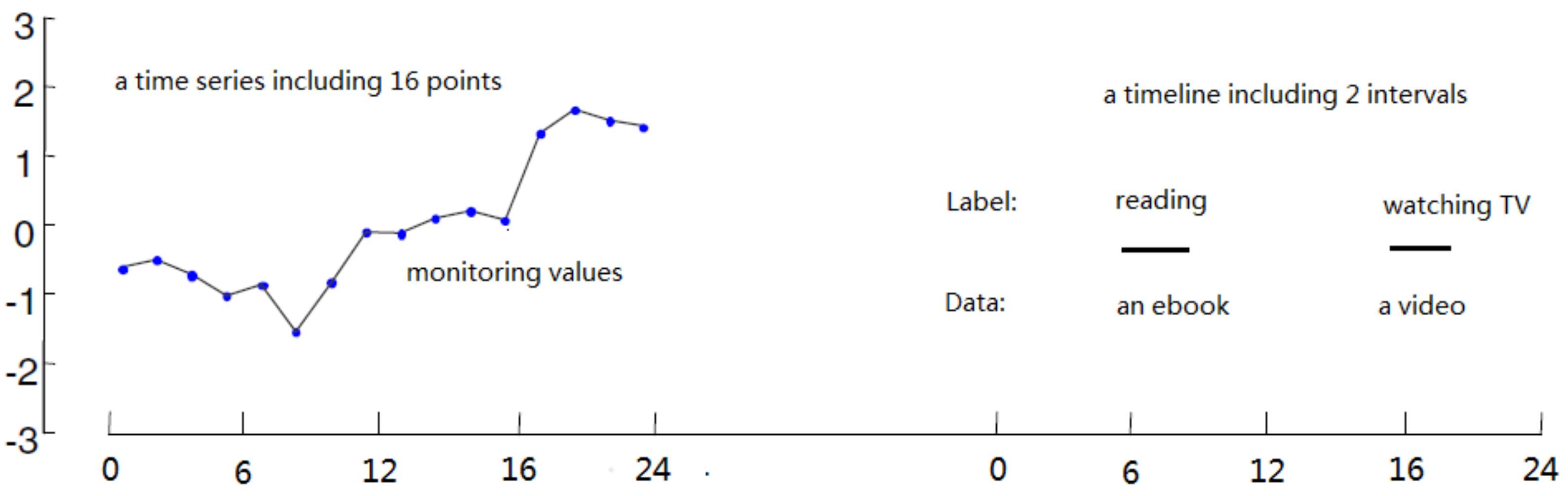
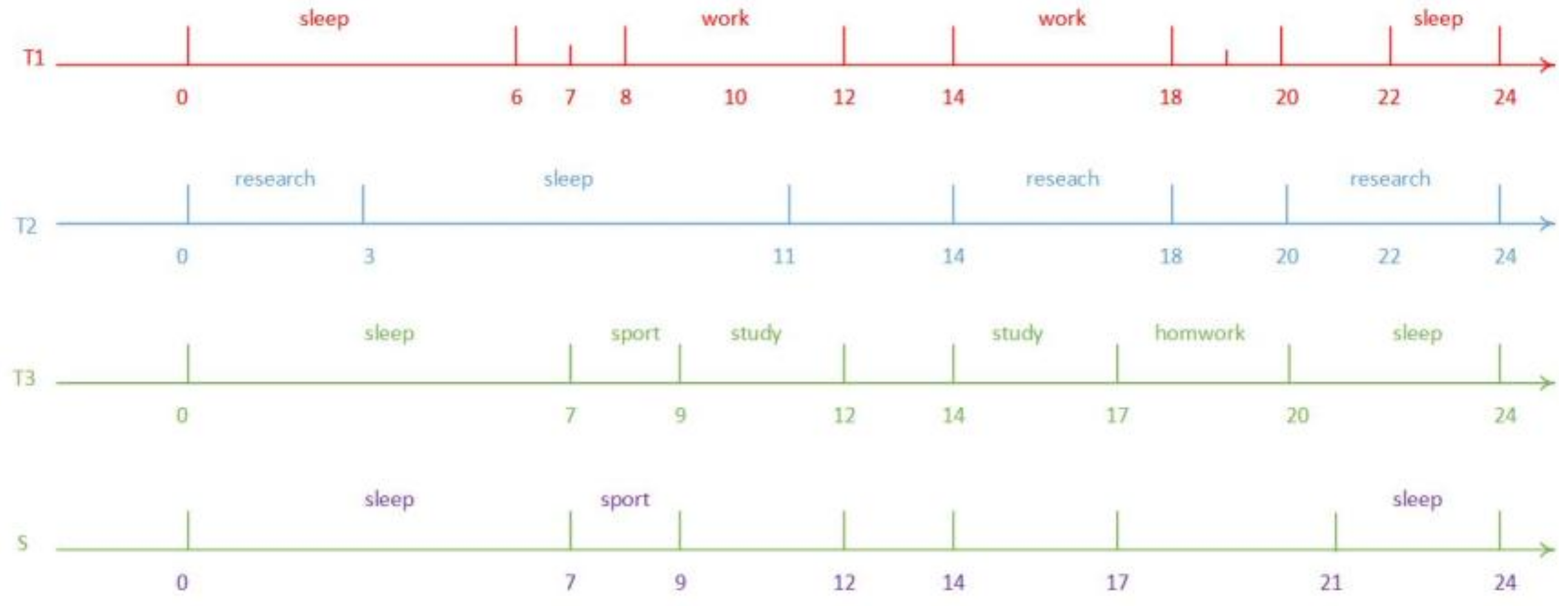

| Interval Type | Expression |
|---|---|
| IT_CLOSED | [Is, Ie] |
| IT_LEFTOPEN | (Is, Ie] |
| IT_RIGHTOPEN | [Is, Ie) |
| IT_OPEN | (Is, Ie) |
| Operator | Definition | Result Shape |
|---|---|---|
| equals | Is = Qs and Ie = Qe | point |
| covers | Is = Qs and Ie = Qe | point, if Q is close and I is open |
| coveredBy | Is = Qs and Ie = Qe | point, if Q is open and I is close |
| Starts | Is = Qs and Qs < Ie < Qe | line |
| startedBy | Is = Qs and Ie > Qe | line |
| Meets | Is ≤ Ie = Qs ≤ Qe | line |
| metBy | Qs ≤ Qe = Is ≤ Ie | line |
| finishes | Qs < Is < Qe and Ie = Qe | line |
| finishedBy | Is < Qs and Ie = Qe | line |
| before | Is ≤ Ie < Qs ≤ Qe | region |
| After | Qs ≤ Qe < Is ≤ Ie | region |
| overlaps | Is < Qs and Qs < Ie < Qe | region |
| overlappedBy | Qs < Is < Qe and Ie>Qe | region |
| during | Qs < Is ≤ Ie < Qe | region |
| contains | Is < Qs ≤ Qe < Ie | region |
| Function | Description |
|---|---|
| cartesian | returns the cartesian product of this timeline and another one, that is, the result of all pairs of elements (a, b) where a is in this and b is in other. |
| count | returns the number of elements in the set. |
| filter | returns a new set containing only the elements that satisfy a predicate. |
| flatMap | returns a new set by first applying a function to all elements of this set, and then flattening the results. |
| foreach | applies a function f to all elements of this set. |
| groupBy | returns a set of grouped elements. |
| intervalize | convert a timeline into a set of intervals |
| min | returns the minimum element from this set as defined by the specified comparator. |
| map | returns a new set by applying a function to all elements of this set. |
| max | returns the maximum element from this set as defined by the specified comparator. |
| reduce | reduces the elements of this set using the specified commutative and associative binary operator. |
| similarity | calculate the similarity of two timelines. |
| Parameters | Description |
|---|---|
| maxTimeValue | the maximum time value of all the timelines generated by generator, the default value is 0. |
| minTimeValue | the minimum time value of all the timelines generated by generator, the default value is 10,000. |
| timelineMinLength | the shortest length of a timeline generated by generator, the default is 10. |
| timelineMaxLength | the longest length of a timeline generated by generator, the default is 50. |
| intervalMinLength | the shortest length of an interval generated by generator, the default is 2. |
| intervalMaxLength | the longest length of an interval generated by generator, the default is 10. |
| intervalAttachment | the range of the attachment data size, the default is [1K, 2K] |
| labelTypes | the maximum types involved in the dataset generated by generator, the default value is 5. |
| numberTimelines | the total number of the timelines generated by generator, the default value is 10,000,000. |
| outputFileName | the generated timeline data file. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Ma, X. A Distributed Indexing Method for Timeline Similarity Query. Algorithms 2018, 11, 41. https://doi.org/10.3390/a11040041
He Z, Ma X. A Distributed Indexing Method for Timeline Similarity Query. Algorithms. 2018; 11(4):41. https://doi.org/10.3390/a11040041
Chicago/Turabian StyleHe, Zhenwen, and Xiaogang Ma. 2018. "A Distributed Indexing Method for Timeline Similarity Query" Algorithms 11, no. 4: 41. https://doi.org/10.3390/a11040041