Rapid Assay to Assess Bacterial Adhesion on Textiles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Method Development
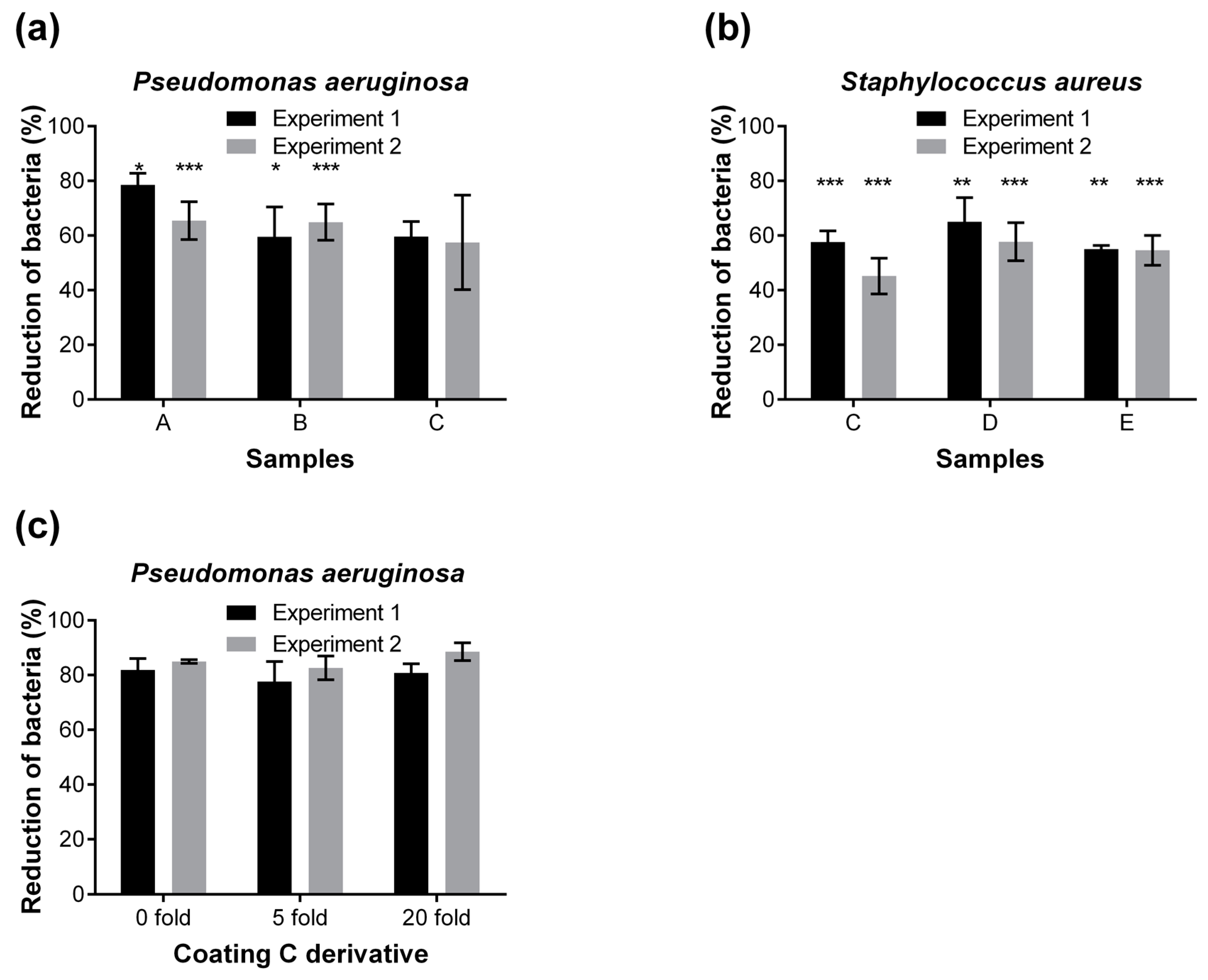
2.2. Screening of Anti-Adhesive Textile Coatings Using the Developed Method
2.3. Identification of Most Promising Anti-Adhesive Textile Coating
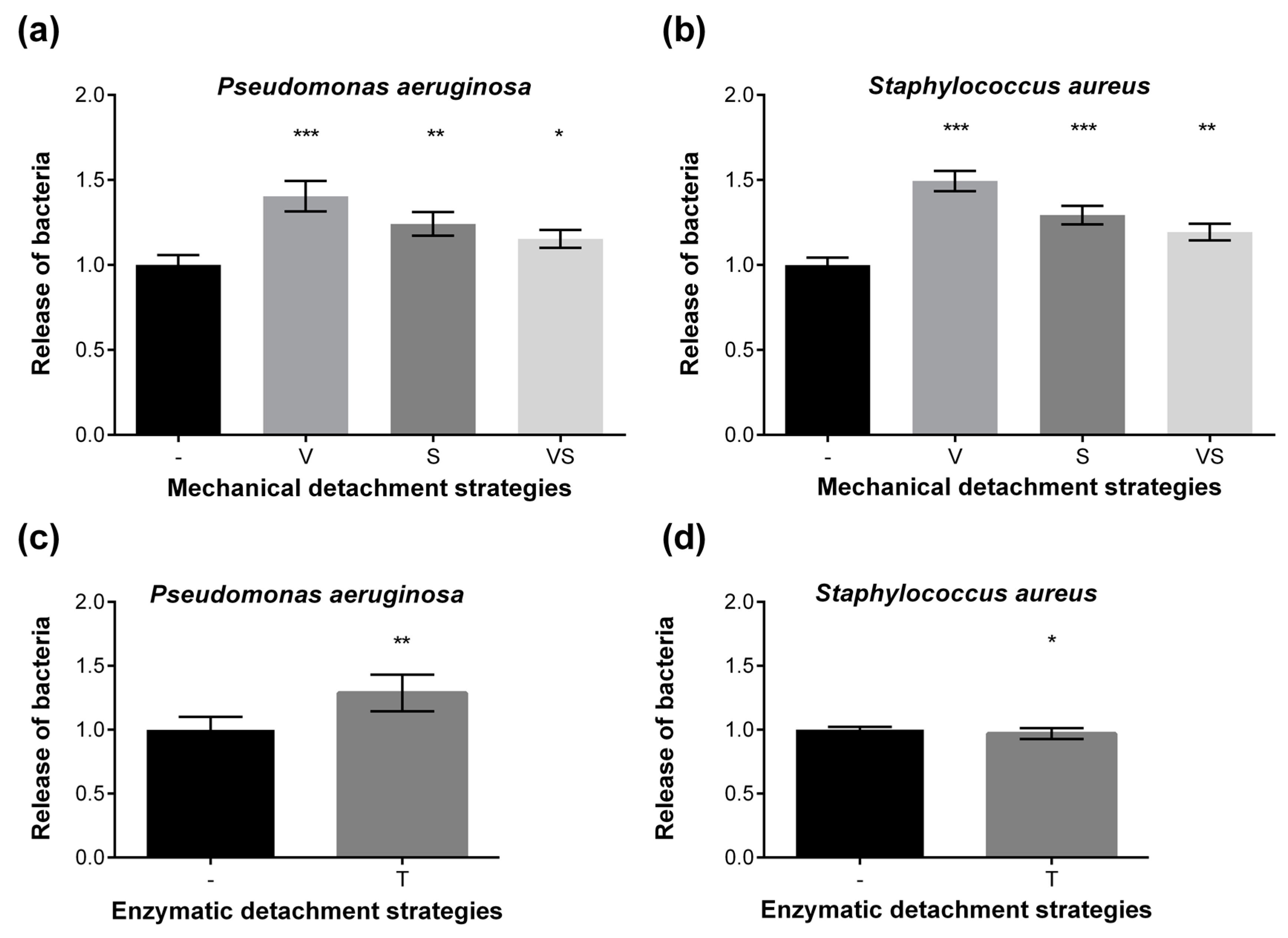
2.4. Washing Stability of Coatings Studied by Bacterial Adhesion Assay and XPS
3. Materials and Methods
3.1. Chemicals and Reagents
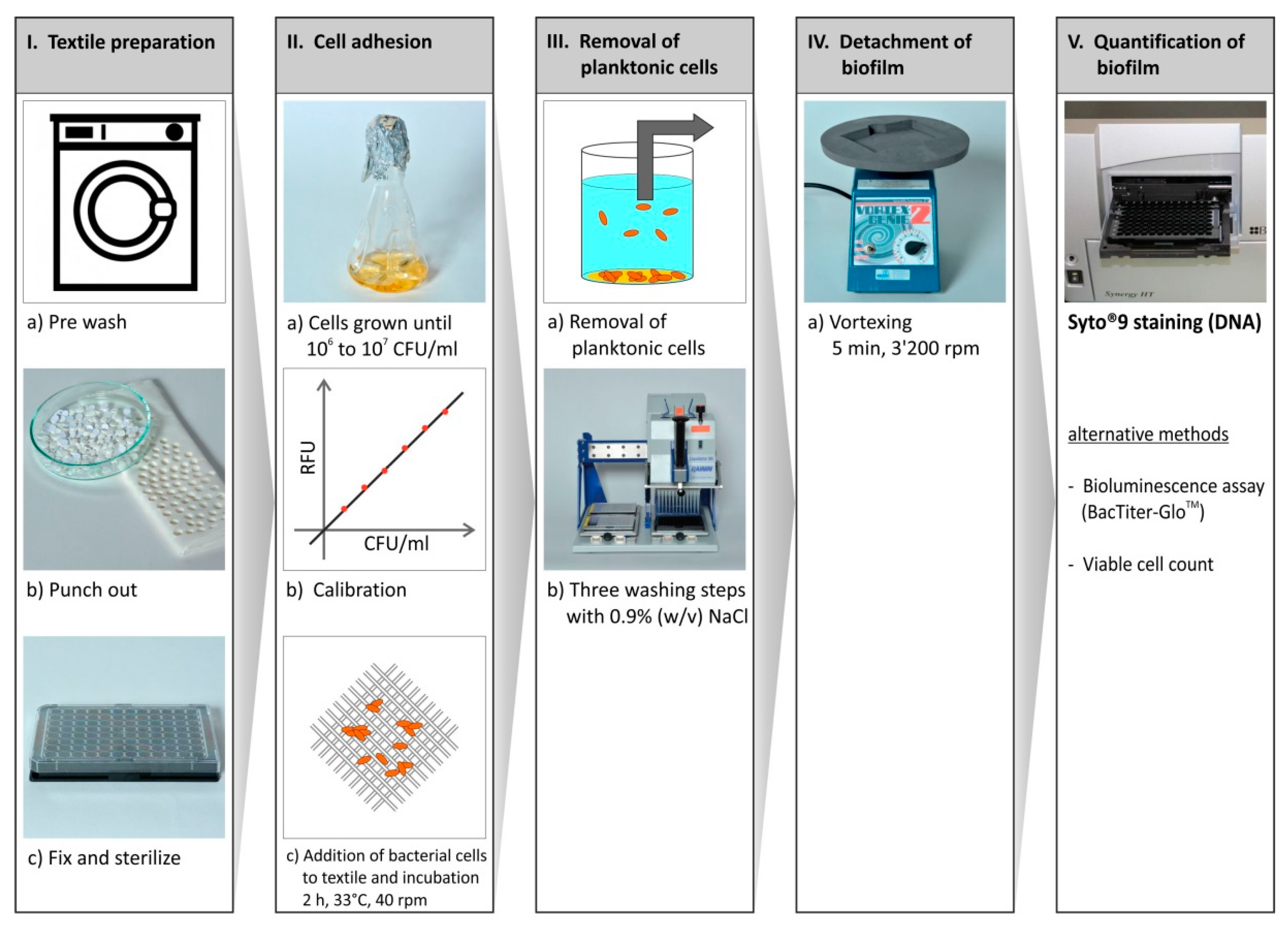
3.2. Bacterial Cultivation and Cell Adhesion Assay
3.3. Quantification of Adhered Bacterial Cells
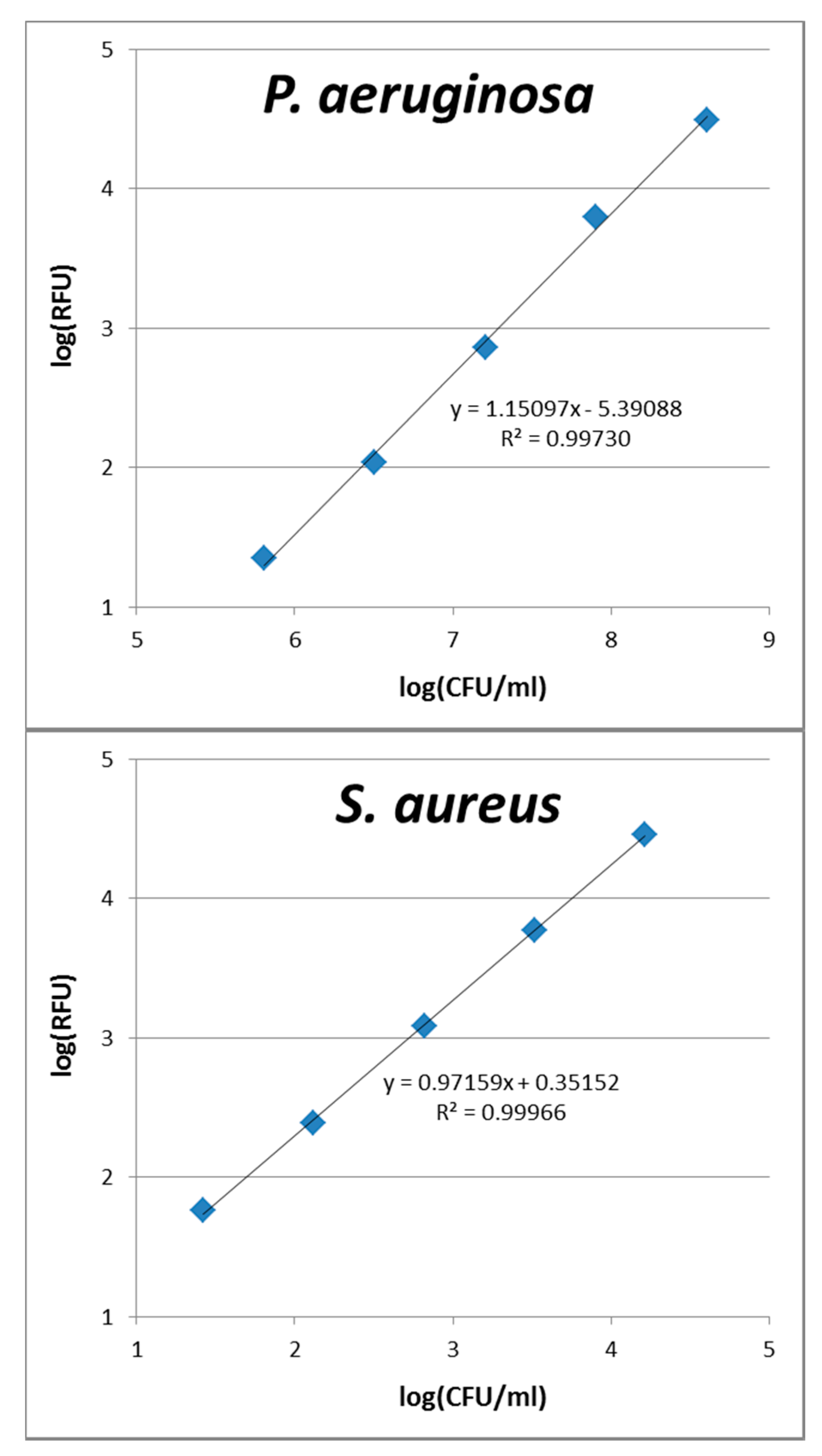
3.4. Calibration of Fluorescence Intensity Versus Cell Numbers
3.5. Washing Resistance Test
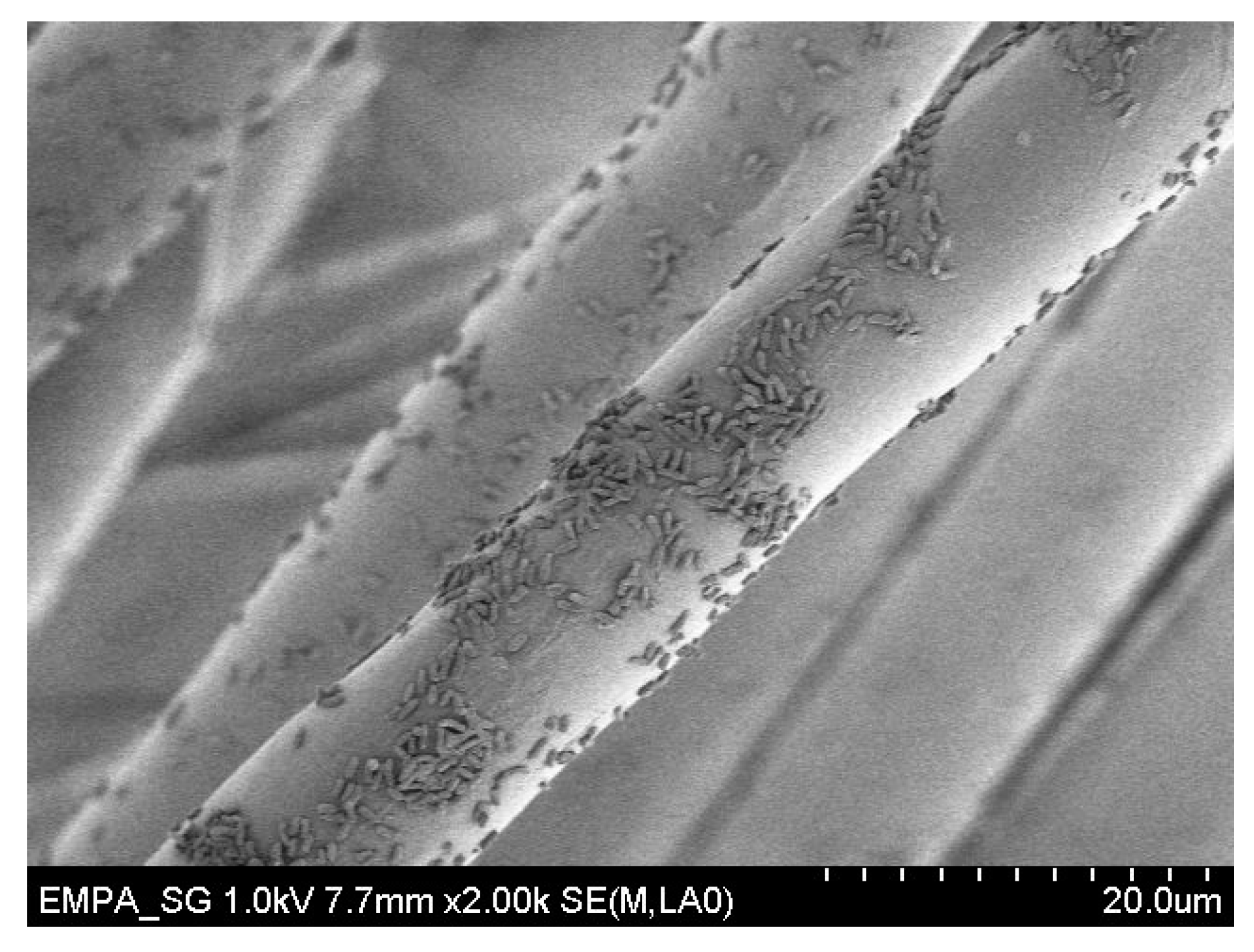
3.6. Scanning Electron Microscopy
3.7. Coating Characterization by X-ray Photoelectron Spectroscopy (XPS)
3.8. Data Analysis for Bacterial Adhesion Tests
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
A.1. Calibration Curves
A.2. Preparation of Fabrics for SEM
A.3. Sample Preparation for SEM Measurements with Hexamethyldisilazane (HMDS)
A.4. Coating Characterization by X-ray Photoelectron Spectroscopy (XPS)
Appendix B
B.1. Characterization of PLL-g-PEG Surfaces via XPS
References
- Szostak-Kotowa, J. Biodeterioration of textiles. Int. Biodeterior. Biodegrad. 2004, 53, 165–170. [Google Scholar] [CrossRef]
- Bajpai, V.; Bajpai, S.; Jha, M.K.; Dey, A.; Ghosh, S. Microbial adherence on textile materials: A review. J. Environ. Res. Dev. 2011, 5, 666–672. [Google Scholar]
- Gao, Y.; Cranston, R. Recent advances in antimicrobial treatments of textiles. Text. Res. J. 2008, 78, 60–72. [Google Scholar]
- Gouveia, I.C. Nanobiotechnology: A new strategy to develop non-toxic antimicrobial textiles for healthcare applications. J. Biotechnol. 2010, 150, 349. [Google Scholar] [CrossRef]
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341–353. [Google Scholar] [CrossRef]
- Wollina, U.; Abdel-Naser, M.B.; Verma, S. Skin physiology and textiles—Consideration of basic interactions. Curr. Probl. Dermatol. 2006, 33, 1–16. [Google Scholar] [PubMed]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 2013, 34, 8533–8554. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, M.; Hakobyan, S.; Ramstedt, M.; Gautrot, J.E. Surface-initiated polymer brushes in the biomedical field: Applications in membrane science, biosensing, cell culture, regenerative medicine and antibacterial coatings. Chem. Rev. 2014, 114, 10976–11026. [Google Scholar] [CrossRef] [PubMed]
- Pinho, E.; Magalhaes, L.; Henriques, M.; Oliveira, R. Antimicrobial activity assessment of textiles: Standard methods comparison. Ann. Microbiol. 2011, 61, 493–498. [Google Scholar] [CrossRef] [Green Version]
- Vigo, T.L.; Benjaminson, M.A. Anti-bacterial fiber treatments and disinfection. Text. Res. J. 1981, 51, 454–465. [Google Scholar] [CrossRef]
- Hsieh, Y.L.; Merry, J. The adherence of Staphylococcus aureus, Staphylococcus epidermidis and Escherichia coli on cotton, polyester, and their blends. J. Appl. Bacteriol. 1986, 60, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.L.; Timm, D.A.; Merry, J. Bacterial adherence on fabrics by a radioisotope labeling method. Text. Res. J. 1987, 57, 20–28. [Google Scholar] [CrossRef]
- McCarthy, B.J. Bioluminescent assay of microbial contamination on textile materials. Int. Biodeterior. Biodegrad. 2001, 48, 79–83. [Google Scholar] [CrossRef]
- Teufel, L.; Schuster, K.C.; Merschak, P.; Bechtold, T.; Redl, B. Development of a fast and reliable method for the assessment of microbial colonization and growth on textiles by DNA quantification. J. Mol. Microbiol. Biotechnol. 2008, 14, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, P.M.; Balaji, S.; Prabhawathi, V.; Neelakandan, R.; Manoharan, P.T.; Doble, M. Effective antibacterial adhesive coating on cotton fabric using ZnO nanorods and chalcone. Carbohydr. Polym. 2010, 79, 717–723. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.G.; Ross, S.A. Ultrasound increases the rate of bacterial cell growth. Biotechnol. Prog. 2003, 19, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Oethinger, M.; Tuohy, M.J.; Procop, G.W.; Bauer, T.W. Improved detection of biofilm-formative bacteria by vortexing and sonication: A pilot study. Clin. Orthop. Relat. Res. 2009, 467, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Kingshott, P.; Wei, J.; Bagge-Ravn, D.; Gadegaard, N.; Gram, L. Covalent attachment of poly(ethylene glycol) to surfaces, critical for reducing bacterial adhesion. Langmuir 2003, 19, 6912–6921. [Google Scholar] [CrossRef]
- Harris, L.G.; Tosatti, S.; Wieland, M.; Textor, M.; Richards, R.G. Staphylococcus aureus adhesion to titanium oxide surfaces coated with non-functionalized and peptide-functionalized poly(l-lysine)-grafted-poly(ethylene glycol) copolymers. Biomaterials 2004, 25, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Maddikeri, R.R.; Tosatti, S.; Schuler, M.; Chessari, S.; Textor, M.; Richards, R.G.; Harris, L.G. Reduced medical infection related bacterial strains adhesion on bioactive rgd modified titanium surfaces: A first step toward cell selective surfaces. J. Biomed. Mater. Res. A 2008, 84, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Tosatti, S.; de Paul, S.M.; Askendal, A.; VandeVondele, S.; Hubbell, J.A.; Tengvall, P.; Textor, M. Peptide functionalized poly(l-lysine)-g-poly(ethylene glycol) on titanium: Resistance to protein adsorption in full heparinized human blood plasma. Biomaterials 2003, 24, 4949–4958. [Google Scholar] [CrossRef]
- Pasche, S.; de Paul, S.M.; Vörös, J.; Spencer, N.D.; Textor, M. Poly(l-lysine)-graft-poly(ethylene glycol) assembled monolayers on niobium oxide surfaces: A quantitative study of the influence of polymer interfacial architecture on resistance to protein adsorption by ToF-SIMS and in situ owls. Langmuir 2003, 19, 9216–9225. [Google Scholar] [CrossRef]
- Ogaki, R.; Andersen, O.Z.; Jensen, G.V.; Kolind, K.; Kraft, D.C.E.; Pedersen, J.S.; Foss, M. Temperature-induced ultradense PEG polyelectrolyte surface grafting provides effective long-term bioresistance against mammalian cells, serum, and whole blood. Biomacromolecules 2012, 13, 3668–3677. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.D.; Ramanathan, V.; Al-Sharif, A.H.; Darouiche, R.O. Efficacy of trypsin in enhancing assessment of bacterial colonisation of vascular catheters. J. Hosp. Infect. 2010, 76, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Moulder, J.; Stickle, W.; Sobol, P.; Bomben, K. Handbook of X-ray Photoelectron Spectroscopy; Perkin-Elmer: Eden Prairie, MN, USA, 1992. [Google Scholar]
- Ivanova, E.P.; Pham, D.K.; Brack, N.; Pigram, P.; Nicolau, D.V. Poly(l-lysine)-mediated immobilisation of oligonucleotides on carboxy-rich polymer surfaces. Biosens. Bioelectron. 2004, 19, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.P.; Michel, R.; Vörös, J.; Textor, M.; Hofer, R.; Rossi, A.; Elbert, D.L.; Hubbell, J.A.; Spencer, N.D. Poly(l-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: Surface-analytical characterization and resistance to serum and fibrinogen adsorption. Langmuir 2001, 17, 489–498. [Google Scholar] [CrossRef]
- Beamson, G.; Briggs, D. High-resolution monochromated X-ray photoelectron-spectroscopy of organic polymers—A comparison between solid-state data for organic polymers and gas-phase data for small molecules. Mol. Phys. 1992, 76, 919–936. [Google Scholar] [CrossRef]
- Amberg, M.; Rupper, P.; Storchenegger, R.; Weder, M.; Hegemann, D. Controlling the release from silver electrodes by titanium adlayers for health monitoring. NanoMedicine 2015, 11, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zürcher, S.; Dorcier, A.; Luengo, G.S.; Spencer, N.D. Adsorption and lubricating properties of poly(l-lysine)-graft-poly(ethylene glycol) on human-hair surfaces. ACS Appl. Mater. Interfaces 2009, 1, 1938–1945. [Google Scholar] [CrossRef] [PubMed]
- Blättler, T.M.; Pasche, S.; Textor, M.; Griesser, H.J. High salt stability and protein resistance of poly(l-lysine)-g-poly(ethylene glycol) copolymers covalently immobilized via aldehyde plasma polymer interlayers on inorganic and polymeric substrates. Langmuir 2006, 22, 5760–5769. [Google Scholar] [CrossRef] [PubMed]
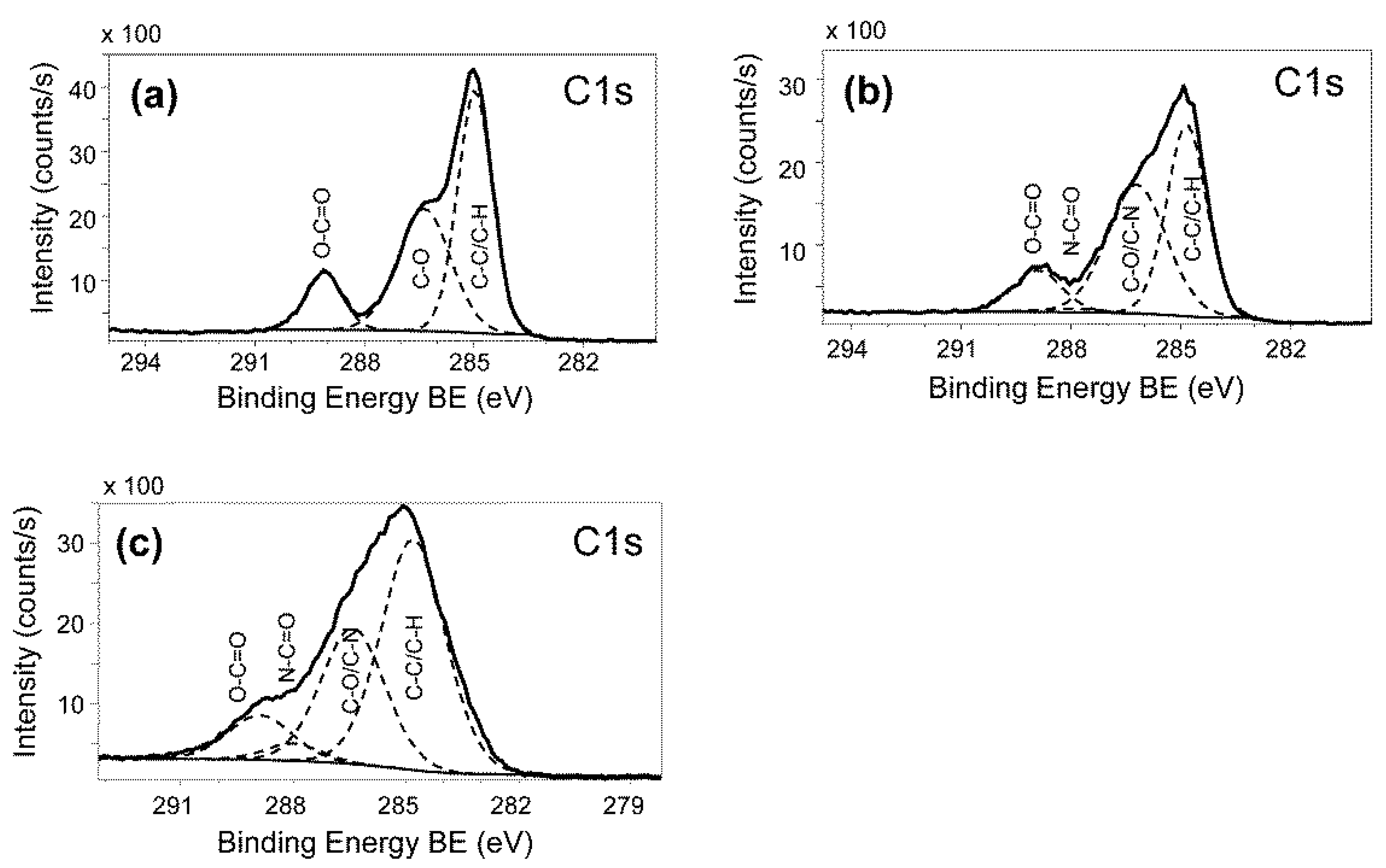
| Sample | % C | % O | % N |
|---|---|---|---|
| PET substratum | 70.9 | 29.1 | 0.0 |
| PLL-g-PEG coated sample | 73.9 | 25.3 | 0.8 |
| coated sample, 20× washed | 73.6 | 25.6 | 0.8 |
| Product Code | Tradename Product | Supplier |
|---|---|---|
| A | Sanitized Research product 1 | MIMOX |
| B | Sanitized Research product 2 | Clariant |
| C | PLL(20)-g[3.5]-PEG(2) | SuSoS |
| D | Cytophob 2 # 121202/3 | PolyAn |
| E | Cytophob 1 # 121201/5 | PolyAn |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt-Emrich, S.; Stiefel, P.; Rupper, P.; Katzenmeier, H.; Amberg, C.; Maniura-Weber, K.; Ren, Q. Rapid Assay to Assess Bacterial Adhesion on Textiles. Materials 2016, 9, 249. https://doi.org/10.3390/ma9040249
Schmidt-Emrich S, Stiefel P, Rupper P, Katzenmeier H, Amberg C, Maniura-Weber K, Ren Q. Rapid Assay to Assess Bacterial Adhesion on Textiles. Materials. 2016; 9(4):249. https://doi.org/10.3390/ma9040249
Chicago/Turabian StyleSchmidt-Emrich, Sabrina, Philipp Stiefel, Patrick Rupper, Heinz Katzenmeier, Caroline Amberg, Katharina Maniura-Weber, and Qun Ren. 2016. "Rapid Assay to Assess Bacterial Adhesion on Textiles" Materials 9, no. 4: 249. https://doi.org/10.3390/ma9040249






