Recent Advances in Scanning Electrochemical Microscopy for Biological Applications
Abstract
:1. Introduction
2. Instrumentation and Operating Modes
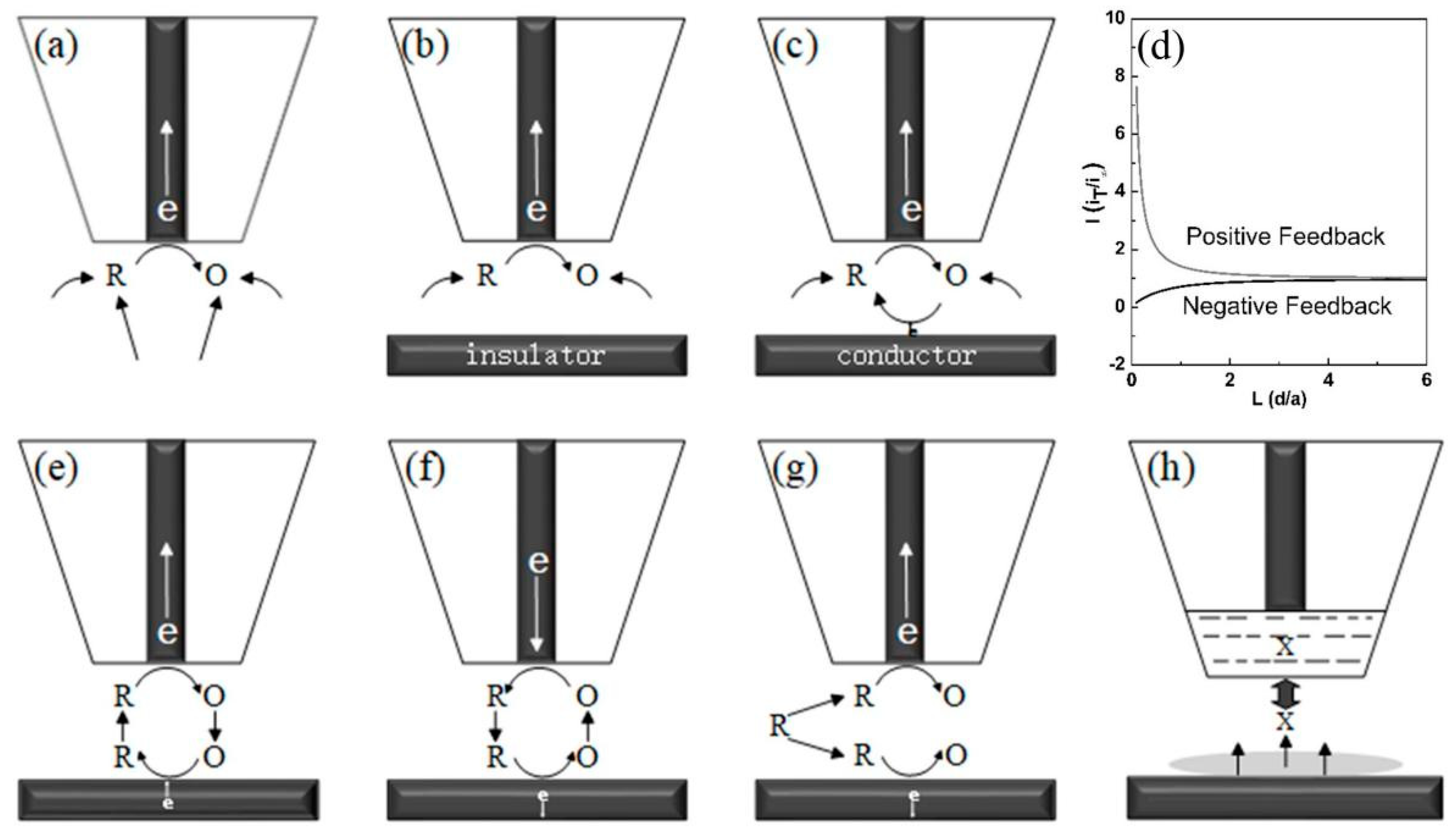
2.1. Feedback Mode
2.2. Generation and Collection Mode
2.3. Redox Competition Mode
2.4. Potentiometric Mode
2.5. Alternating Current Mode
2.6. Penetration Mode
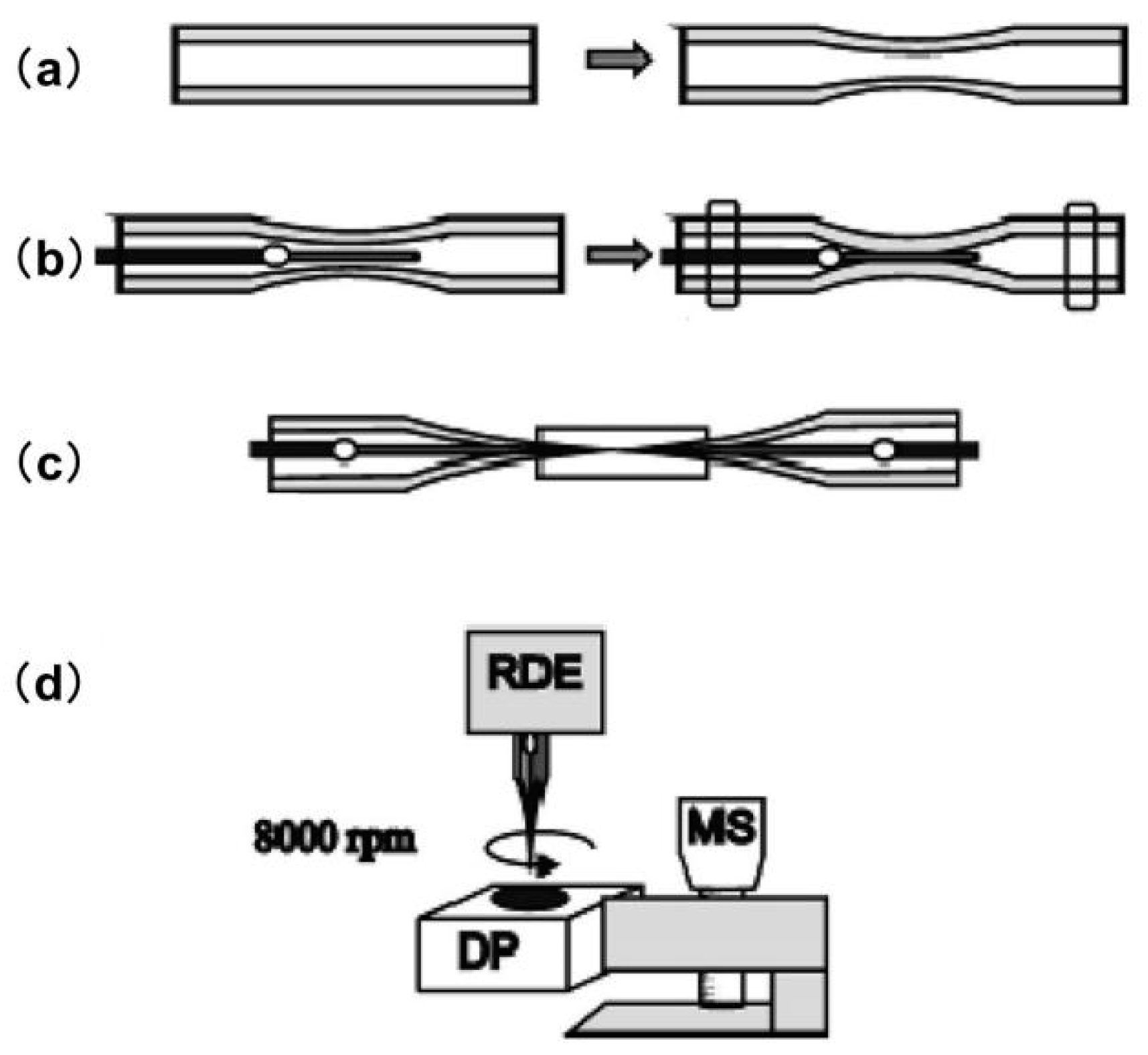
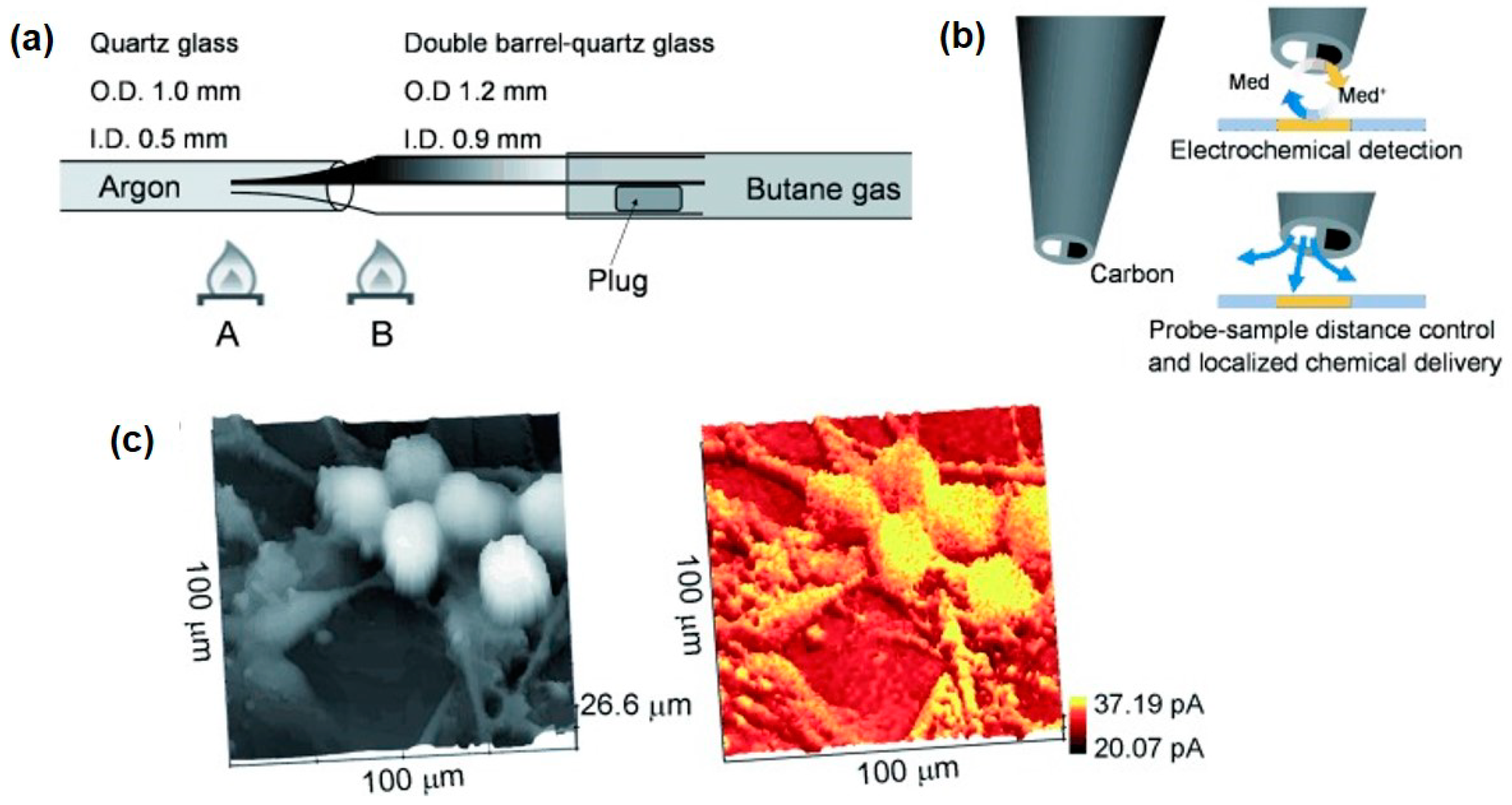
2.7. Probes
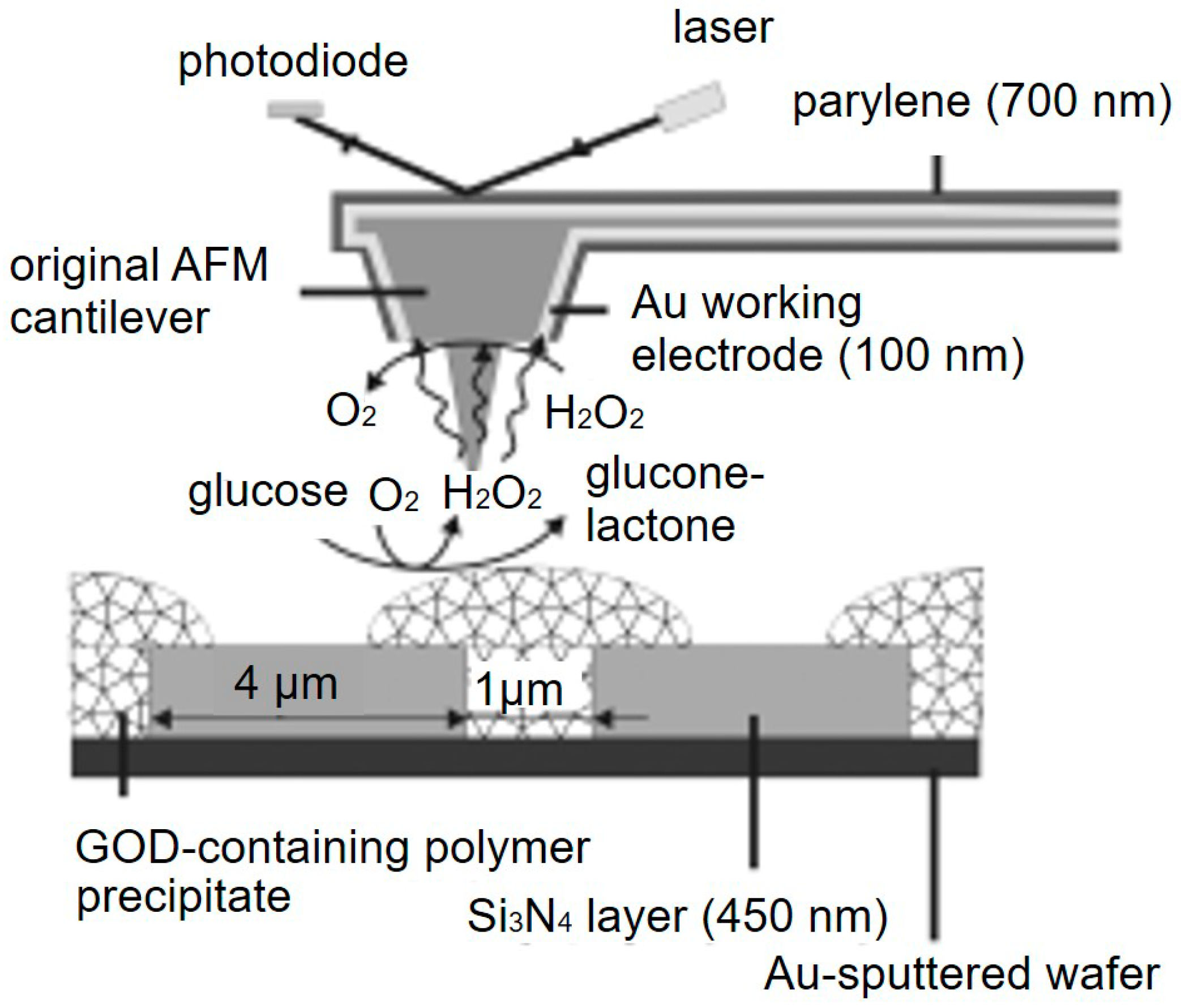
2.8. Multi-Sensing Platform
2.9. Modeling for SECM Modes
3. Applications
3.1. Imaging of Enzyme Activity
3.2. Detection of DNA or Hybridization
3.3. Investigation of Protein Activity
3.4. Living Cell Studies
3.5. Study of Bacterial Behavior and Biofilm Activity
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bard, A.J.; Fan, F.R.F.; Kwak, J.; Lev, O. Scanning electrochemical microscopy. Introduction and principles. Anal. Chem. 1989, 61, 132–138. [Google Scholar] [CrossRef]
- Liu, H.Y.; Fan, F.R.F.; Lin, C.W.; Bard, A.J. Scanning electrochemical and tunneling ultramicroelectrode microscope for high-resolution examination of electrode surfaces in solution. J. Am. Chem. Soc. 1986, 108, 3838–3839. [Google Scholar] [CrossRef]
- Engstrom, R.C.; Weber, M.; Wunder, D.J.; Burgess, R.; Winquist, S. Measurements within the diffusion layer using a microelectrode probe. Anal. Chem. 1986, 58, 844–848. [Google Scholar] [CrossRef]
- Engstrom, R.C.; Meaney, T.; Tople, R.; Wightman, R.M. Spatiotemporal description of the diffusion layer with a microelectrode probe. Anal. Chem. 1987, 59, 2005–2010. [Google Scholar] [CrossRef]
- Kwak, J.; Bard, A.J. Scanning electrochemical microscopy. Theory of the feedback mode. Anal. Chem. 1989, 61, 1221–1227. [Google Scholar] [CrossRef]
- Lee, C.; Kwak, J.; Bard, A.J. Application of scanning electrochemical microscopy to biological samples. Proc. Natl. Acad. Sci. USA 1990, 87, 1740–1743. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.A.; Martin, S.; Whitworth, A.L.; Macpherson, J.V.; Unwin, P.R. Scanning electrochemical microscopy: Principles and applications to biophysical systems. Physiol. Meas. 2006, 27, R63–R108. [Google Scholar] [CrossRef] [PubMed]
- Bard, A.J.; Li, X.; Zhan, W. Chemically imaging living cells by scanning electrochemical microscopy. Biosens. Bioelectron. 2006, 22, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, I.; Kuss, S.; Mauzeroll, J.; Geissler, M. Biological Scanning Electrochemical Microscopy and Its Application to Live Cell Studies. Anal. Chem. 2011, 83, 1485–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergner, S.; Vatsyayan, P.; Matysik, F.-M. Recent advances in high resolution scanning electrochemical microscopy of living cells—A review. Anal. Chim. Acta 2013, 775, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.; Nagy, G. Application of Scanning Electrochemical Microscopy in Bioanalytical Chemistry. In Trends in Bioelectroanalysis; Matysik, F.-M., Ed.; Springer: Cham, Switzerland, 2016; Volume 6, pp. 281–339. ISBN 978-3-319-48483-9. [Google Scholar]
- Zoski, C.G. Review—Advances in Scanning Electrochemical Microscopy (SECM). J. Electrochem. Soc. 2016, 163, H3088–H3100. [Google Scholar] [CrossRef]
- Polcari, D.; Dauphin-Ducharme, P.; Mauzeroll, J. Scanning Electrochemical Microscopy: A Comprehensive Review of Experimental Parameters from 1989 to 2015. Chem. Rev. 2016, 116, 13234–13278. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y. Development of High-Resolution Scanning Electrochemical Microscopy for Nanoscale Topography and Electrochemical Simultaneous Imaging. Electrochemistry 2016, 84, 662–666. [Google Scholar] [CrossRef] [Green Version]
- Li, M.S.M.; Filice, F.P.; Ding, Z. Determining live cell topography by scanning electrochemical microscopy. J. Electroanal. Chem. 2016, 779, 176–186. [Google Scholar] [CrossRef]
- Mauzeroll, J.; Bard, A.J. Scanning electrochemical microscopy of menadione-glutathione conjugate export from yeast cells. Proc. Natl. Acad. Sci. USA 2004, 101, 7862–7867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauzeroll, J.; Bard, A.J.; Owhadian, O.; Monks, T.J. Menadione metabolism to thiodione in hepatoblastoma by scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2004, 101, 17582–17587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xiao, X.; Su, T.; Gu, X.; Jin, T.; Du, L.; Tang, J. A novel thermocouple microelectrode for applications in SECM and variable temperature electrochemistry. Electrochem. Commun. 2014, 47, 71–74. [Google Scholar] [CrossRef]
- Barforoush, J.M.; McDonald, T.D.; Desai, T.A.; Widrig, D.; Bayer, C.; Brown, M.K.; Cummings, L.C.; Leonard, K.C. Intelligent Scanning Electrochemical Microscopy Tip and Substrate Control Utilizing Fuzzy Logic. Electrochimica Acta 2016, 190, 713–719. [Google Scholar] [CrossRef]
- Kim, J.; Shen, M.; Nioradze, N.; Amemiya, S. Stabilizing Nanometer Scale Tip-to-Substrate Gaps in Scanning Electrochemical Microscopy Using an Isothermal Chamber for Thermal Drift Suppression. Anal. Chem. 2012, 84, 3489–3492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Renault, C.; Nioradze, N.; Arroyo-Currás, N.; Leonard, K.C.; Bard, A.J. Electrocatalytic Activity of Individual Pt Nanoparticles Studied by Nanoscale Scanning Electrochemical Microscopy. J. Am. Chem. Soc. 2016, 138, 8560–8568. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Renault, C.; Nioradze, N.; Arroyo-Currás, N.; Leonard, K.C.; Bard, A.J. Nanometer Scale Scanning Electrochemical Microscopy Instrumentation. Anal. Chem. 2016, 88, 10284–10289. [Google Scholar] [CrossRef] [PubMed]
- Koley, D.; Bard, A.J. Triton X-100 concentration effects on membrane permeability of a single HeLa cell by scanning electrochemical microscopy (SECM). Proc. Natl. Acad. Sci. USA 2010, 107, 16783–16787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, Z.; Topping, K.; Dong, B.; Shamsi, M.H.; Kraatz, H.B. An unexpected use of ferrocene. A scanning electrochemical microscopy study of a toll-like receptor array and its interaction with E. coli. Chem. Commun. 2017, 53, 2946–2949. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; She, Z.; Lin, Y.-C.; Martić, S.; Mann, D.J.; Kraatz, H.-B. Clickable 5′-γ-Ferrocenyl Adenosine Triphosphate Bioconjugates in Kinase-Catalyzed Phosphorylations. Chem.-Eur. J. 2015, 21, 4988–4999. [Google Scholar] [CrossRef] [PubMed]
- Mureşan, L.; Nistor, M.; Gáspár, S.; Popescu, I.C.; Csöregi, E. Monitoring of glucose and glutamate using enzyme microstructures and scanning electrochemical microscopy. Bioelectrochemistry 2009, 76, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Bard, A.J.; Mirkin, M.V. Scanning Electrochemical Microscopy; CRC Press: Boca Raton, FL, USA, 2012; ISBN 978-1-4398-3113-7. [Google Scholar]
- Huang, C.C.; Wang, Q.; Xiang, D.B.; Shao, H.B. Surface interrogation mode of scanning electrochemical microscopy for oxidation study of adsorbed CO generated from serine on platinum. Chin. Chem. Lett. 2011, 22, 1481–1484. [Google Scholar] [CrossRef]
- Hirano, Y.; Kowata, K.; Kodama, M.; Komatsu, Y. Development of a scanning electrochemical microscopy-based micropipette and its application to analysis of topographic change of single-cell. Bioelectrochemistry 2013, 92, 1–5. [Google Scholar] [CrossRef] [PubMed]
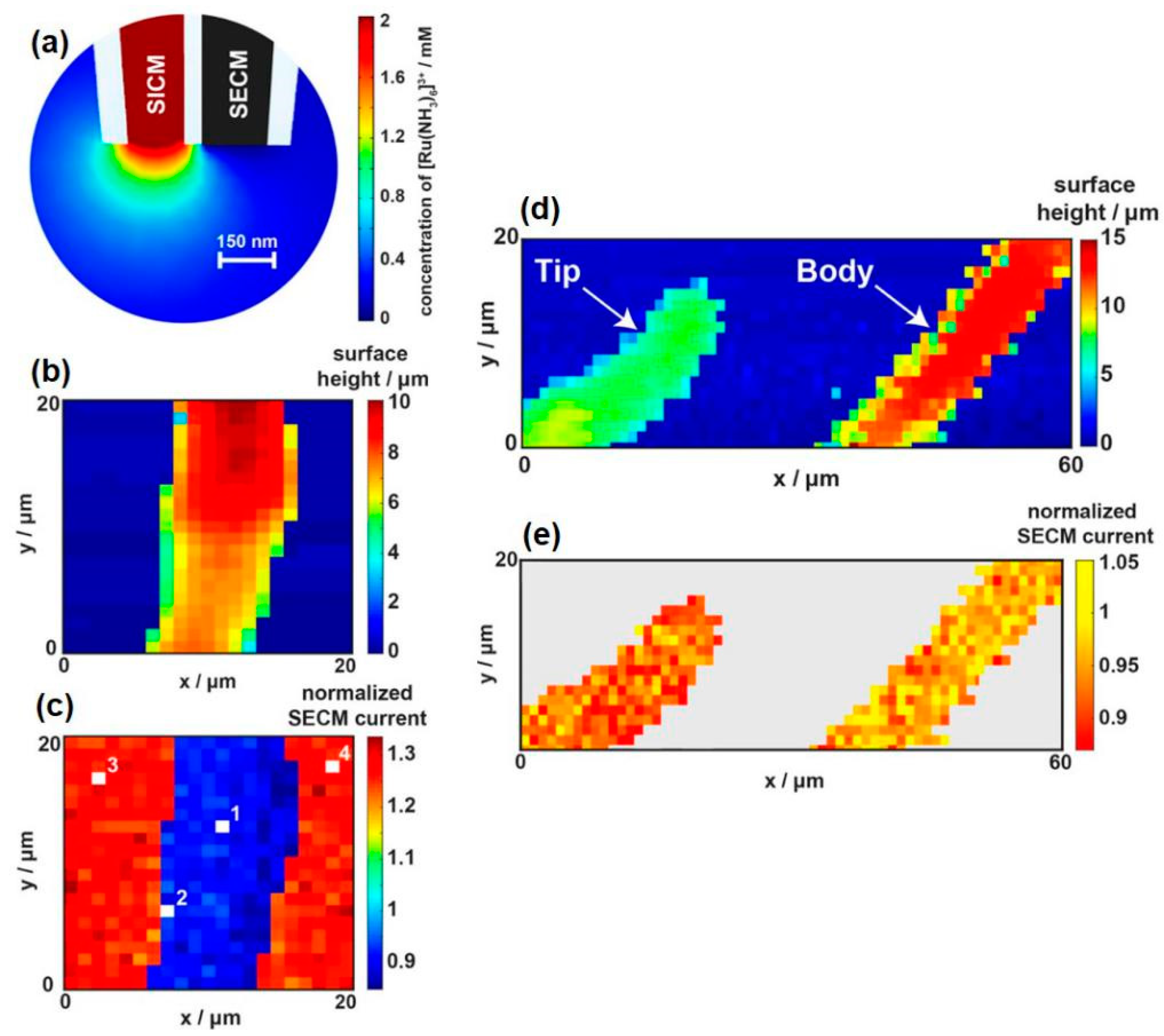
- Page, A.; Kang, M.; Armitstead, A.; Perry, D.; Unwin, P.R. Quantitative Visualization of Molecular Delivery and Uptake at Living Cells with Self-Referencing Scanning Ion Conductance Microscopy-Scanning Electrochemical Microscopy. Anal. Chem. 2017, 89, 3021–3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckhard, K.; Chen, X.; Turcu, F.; Schuhmann, W. Redox competition mode of scanning electrochemical microscopy (RC-SECM) for visualisation of local catalytic activity. Phys. Chem. Chem. Phys. 2006, 8, 5359–5365. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Deng, L.; Zhang, D.; Qian, H.; Du, C.; Li, X.; Mol, J.M.C.; Terryn, H.A. Shape memory composite (SMC) self-healing coatings for corrosion protection. Prog. Org. Coat. 2016, 97, 261–268. [Google Scholar] [CrossRef]
- Qian, H.; Xu, D.; Du, C.; Zhang, D.; Li, X.; Huang, L.; Deng, L.; Tu, Y.; Mol, J.M.C.; Terryn, H.A. Dual-action smart coatings with a self-healing superhydrophobic surface and anti-corrosion properties. J. Mater. Chem. A 2017, 5, 2355–2364. [Google Scholar] [CrossRef] [Green Version]
- Karnicka, K.; Eckhard, K.; Guschin, D.A.; Stoica, L.; Kulesza, P.J.; Schuhmann, W. Visualisation of the local bio-electrocatalytic activity in biofuel cell cathodes by means of redox competition scanning electrochemical microscopy (RC-SECM). Electrochem. Commun. 2007, 9, 1998–2002. [Google Scholar] [CrossRef]
- Ivanauskas, F.; Morkvenaite-Vilkonciene, I.; Astrauskas, R.; Ramanavicius, A. Modelling of Scanning Electrochemical Microscopy at Redox Competition Mode Using Diffusion and Reaction Equations. Electrochim. Acta 2016, 222, 347–354. [Google Scholar] [CrossRef]
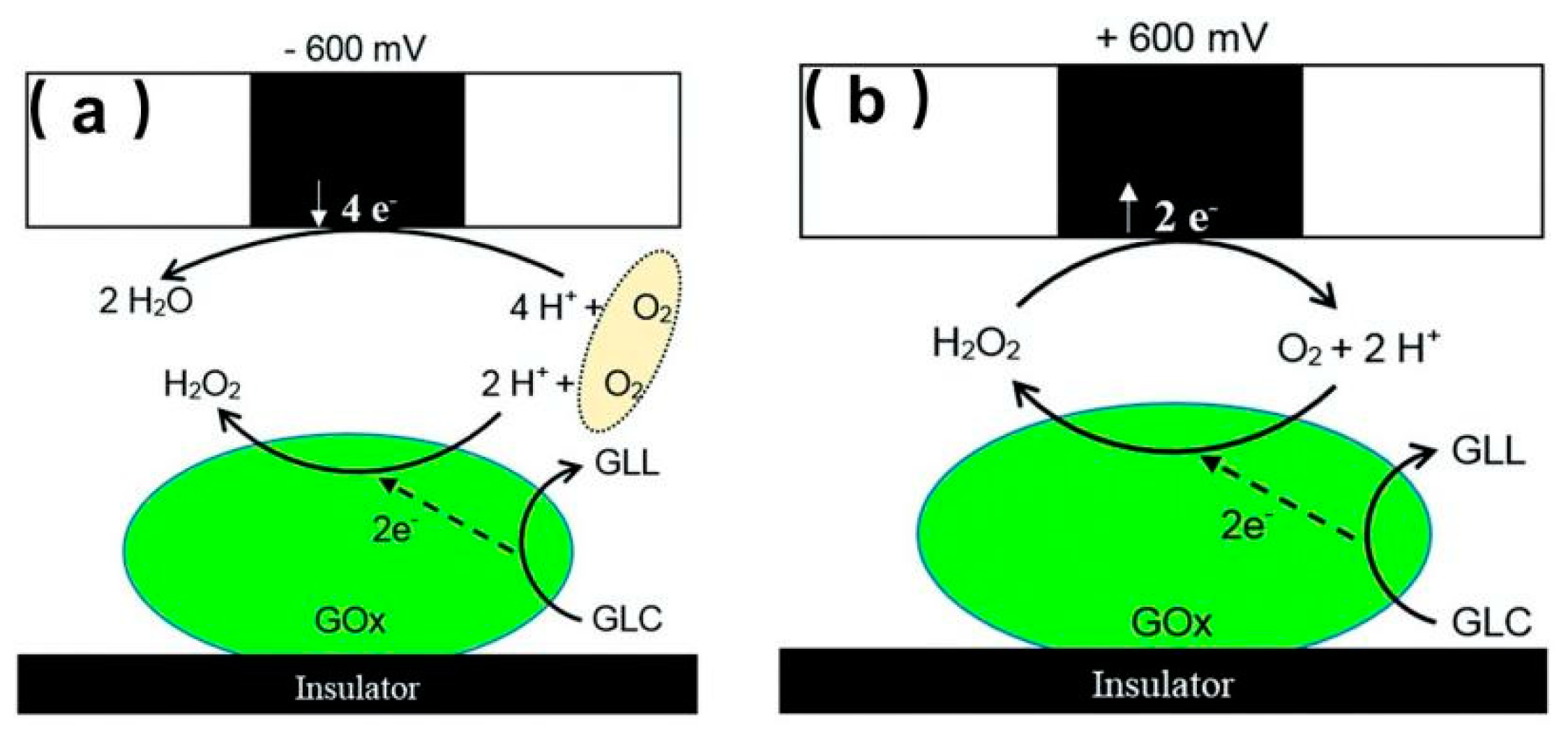
- Morkvenaite-Vilkonciene, I.; Ramanaviciene, A.; Genys, P.; Ramanavicius, A. Evaluation of Enzymatic Kinetics of GOx-based Electrodes by Scanning Electrochemical Microscopy at Redox Competition Mode. Electroanalysis 2017, 29, 1532–1542. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, X.; Ye, Z.; Zhang, J.; Cao, F.; Zhang, J. A fabrication of iridium oxide film pH micro-sensor on Pt ultramicroelectrode and its application on in-situ pH distribution of 316L stainless steel corrosion at open circuit potential. Sens. Actuators B Chem. 2018, 255, 1974–1982. [Google Scholar] [CrossRef]
- Horrocks, B.R.; Mirkin, M.V.; Pierce, D.T.; Bard, A.J.; Nagy, G.; Toth, K. Scanning electrochemical microscopy. 19. Ion-selective potentiometric microscopy. Anal. Chem. 1993, 65, 1213–1224. [Google Scholar] [CrossRef]
- Cornut, R.; Lefrou, C. A unified new analytical approximation for negative feedback currents with a microdisk SECM tip. J. Electroanal. Chem. 2007, 608, 59–66. [Google Scholar] [CrossRef]
- Zhu, Z.; Ye, Z.; Zhang, Q.; Zhang, J.; Cao, F. Novel dual Pt-Pt/IrO x ultramicroelectrode for pH imaging using SECM in both potentiometric and amperometric modes. Electrochem. Commun. 2018, 88, 47–51. [Google Scholar] [CrossRef]
- Kiss, A.; Nagy, G. New SECM scanning algorithms for improved potentiometric imaging of circularly symmetric targets. Electrochimica Acta 2014, 119, 169–174. [Google Scholar] [CrossRef]
- Kiss, A.; Nagy, G. Deconvolution of potentiometric SECM images recorded with high scan rate. Electrochimica Acta 2015, 163, 303–309. [Google Scholar] [CrossRef]
- Diakowski, P.M.; Baranski, A.S. Positive and negative AC impedance feedback observed above conductive substrates under SECM conditions. Electrochim. Acta 2006, 52, 854–862. [Google Scholar] [CrossRef]
- Zhao, S.H.; Zheng, J.Y.; Yang, J.Y.; Lin, C.G.; Wang, W.; Qiu, R.; Chen, Q. The Alternating Current Scanning Electrochemical Microscopy Characterization of Diatom Cell Activity Affected by Paeonol. Appl. Mech. Mater. 2014, 707, 113–116. [Google Scholar] [CrossRef]
- Diakowski, P.M.; Ding, Z. Interrogation of living cells using alternating current scanning electrochemical microscopy (AC-SECM). Phys. Chem. Chem. Phys. 2007, 9, 5966–5974. [Google Scholar] [CrossRef] [PubMed]
- Morkvenaite-Vilkonciene, I.; Genys, P.; Ramanaviciene, A.; Ramanavicius, A. Scanning electrochemical impedance microscopy for investigation of glucose oxidase catalyzed reaction. Colloids Surf. B Biointerfaces 2015, 126, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; Tokar, S.; Clausmeyer, J.; Babakinejad, B.; Mikhaleva, S.; Cornut, R.; Takahashi, Y.; López Córdoba, A.; Novak, P.; Shevchuck, A.I.; et al. Electrochemical Nanoprobes for Single-Cell Analysis. ACS Nano 2014, 8, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrlau, M.G.; Dun, N.J.; Bau, H.H. Cell Electrophysiology with Carbon Nanopipettes. ACS Nano 2009, 3, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Miller, C.J.; Bard, A.J. Scanning electrochemical microscopy: Preparation of submicrometer electrodes. Anal. Chem. 1991, 63, 78–83. [Google Scholar] [CrossRef]
- Katemann, B.B.; Schuhmann, W. Fabrication and Characterization of Needle-Type Pt-Disk Nanoelectrodes. Electroanalysis 2002, 14, 22–28. [Google Scholar] [CrossRef]
- Li, M.S.M.; Filice, F.P.; Ding, Z. Scanning electrochemical microscopy imaging with laser-pulled probes. J. Electroanal. Chem. 2016, 781, 126–135. [Google Scholar] [CrossRef]
- Cao, F.; Kim, J.; Bard, A.J. Detection of the Short-Lived Cation Radical Intermediate in the Electrochemical Oxidation of N,N-Dimethylaniline by Scanning Electrochemical Microscopy. J. Am. Chem. Soc. 2014, 136, 18163–18169. [Google Scholar] [CrossRef] [PubMed]
- Heinze, J. Ultramicroelectrodes in electrochemistry. Angew. Chem. Int. Ed. Engl. 1993, 32, 1268–1288. [Google Scholar] [CrossRef]
- Danis, L.; Polcari, D.; Kwan, A.; Gateman, S.M.; Mauzeroll, J. Fabrication of Carbon, Gold, Platinum, Silver, and Mercury Ultramicroelectrodes with Controlled Geometry. Anal. Chem. 2015, 87, 2565–2569. [Google Scholar] [CrossRef] [PubMed]
- Clausmeyer, J.; Schuhmann, W. Nanoelectrodes: Applications in electrocatalysis, single-cell analysis and high-resolution electrochemical imaging. TrAC Trends Anal. Chem. 2016, 79, 46–59. [Google Scholar] [CrossRef]
- Fan, Y.; Han, C.; Zhang, B. Recent advances in the development and application of nanoelectrodes. Analyst 2016, 141, 5474–5487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Zhang, Z.; Guo, J.; Shao, Y. Fabrication of Nanometer-Sized Electrodes and Tips for Scanning Electrochemical Microscopy. Anal. Chem. 2001, 73, 5346–5351. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Kucernak, A. Fabrication of carbon microelectrodes with an effective radius of 1 nm. Electrochem. Commun. 2002, 4, 80–85. [Google Scholar] [CrossRef]
- Mezour, M.A.; Morin, M.; Mauzeroll, J. Fabrication and Characterization of Laser Pulled Platinum Microelectrodes with Controlled Geometry. Anal. Chem. 2011, 83, 2378–2382. [Google Scholar] [CrossRef] [PubMed]
- Liebetrau, J.M.; Miller, H.M.; Baur, J.E.; Takacs, S.A.; Anupunpisit, V.; Garris, P.A.; Wipf, D.O. Scanning Electrochemical Microscopy of Model Neurons: Imaging and Real-Time Detection of Morphological Changes. Anal. Chem. 2003, 75, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Bauermann, L.P.; Schuhmann, W.; Schulte, A. An advanced biological scanning electrochemical microscope (Bio-SECM) for studying individual living cells. Phys. Chem. Chem. Phys. 2004, 6, 4003–4008. [Google Scholar] [CrossRef]
- Ludwig, M.; Kranz, C.; Schuhmann, W.; Gaub, H.E. Topography feedback mechanism for the scanning electrochemical microscope based on hydrodynamic forces between tip and sample. Rev. Sci. Instrum. 1995, 66, 2857–2860. [Google Scholar] [CrossRef]
- Ballesteros Katemann, B.; Schulte, A.; Schuhmann, W. Constant-Distance Mode Scanning Electrochemical Microscopy (SECM)—Part I: Adaptation of a Non-Optical Shear-Force-Based Positioning Mode for SECM Tips. Chem.-Eur. J. 2003, 9, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- Nebel, M.; Eckhard, K.; Erichsen, T.; Schulte, A.; Schuhmann, W. 4D Shearforce-Based Constant-Distance Mode Scanning Electrochemical Microscopy. Anal. Chem. 2010, 82, 7842–7848. [Google Scholar] [CrossRef] [PubMed]
- Danis, L.; Snowden, M.E.; Tefashe, U.M.; Heinemann, C.N.; Mauzeroll, J. Development of Nano-Disc electrodes for Application as Shear Force Sensitive Electrochemical Probes. Electrochim. Acta 2014, 136, 121–129. [Google Scholar] [CrossRef]
- Nebel, M.; Grützke, S.; Diab, N.; Schulte, A.; Schuhmann, W. Visualization of Oxygen Consumption of Single Living Cells by Scanning Electrochemical Microscopy: The Influence of the Faradaic Tip Reaction. Angew. Chem. Int. Ed. 2013, 52, 6335–6338. [Google Scholar] [CrossRef] [PubMed]
- Northcutt, R.; Maddox, J.; Sundaresan, V.-B. Electrode Fabrication for Scanning Electrochemical Microscopy and Shear Force Imaging. In ASME 2016 Conference on Smart Materials, Adaptive Structures and Intelligent Systems; American Society of Mechanical Engineers: New York, NY, USA, 2016; p. V002T06A013. [Google Scholar]
- Kranz, C. Recent advancements in nanoelectrodes and nanopipettes used in combined scanning electrochemical microscopy techniques. Analyst 2014, 139, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.A.; Fernández, J.L.; Mauzeroll, J.; Bard, A.J. Scanning Electrochemical Microscopy. 55. Fabrication and Characterization of Micropipet Probes. Anal. Chem. 2005, 77, 5182–5188. [Google Scholar] [CrossRef] [PubMed]
- Comstock, D.J.; Elam, J.W.; Pellin, M.J.; Hersam, M.C. Integrated Ultramicroelectrode−Nanopipet Probe for Concurrent Scanning Electrochemical Microscopy and Scanning Ion Conductance Microscopy. Anal. Chem. 2010, 82, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shevchuk, A.I.; Novak, P.; Zhang, Y.; Ebejer, N.; Macpherson, J.V.; Unwin, P.R.; Pollard, A.J.; Roy, D.; Clifford, C.A.; et al. Multifunctional Nanoprobes for Nanoscale Chemical Imaging and Localized Chemical Delivery at Surfaces and Interfaces. Angew. Chem. Int. Ed. 2011, 50, 9638–9642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Shevchuk, A.I.; Novak, P.; Babakinejad, B.; Macpherson, J.; Unwin, P.R.; Shiku, H.; Gorelik, J.; Klenerman, D.; Korchev, Y.E.; et al. Topographical and electrochemical nanoscale imaging of living cells using voltage-switching mode scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 11540–11545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macpherson, J.V.; Unwin, P.R. Combined Scanning Electrochemical−Atomic Force Microscopy. Anal. Chem. 2000, 72, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.D.; Anne, A.; Cambril, E.; Demaille, C. Optimized hand fabricated AFM probes for simultaneous topographical and electrochemical tapping mode imaging. Ultramicroscopy 2011, 111, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Dobson, P.S.; Weaver, J.M.R.; Holder, M.N.; Unwin, P.R.; Macpherson, J.V. Characterization of Batch-Microfabricated Scanning Electrochemical-Atomic Force Microscopy Probes. Anal. Chem. 2005, 77, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Eifert, A.; Mizaikoff, B.; Kranz, C. Advanced fabrication process for combined atomic force-scanning electrochemical microscopy (AFM-SECM) probes. Micron 2015, 68, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Nault, L.; Taofifenua, C.; Anne, A.; Chovin, A.; Demaille, C.; Besong-Ndika, J.; Cardinale, D.; Carette, N.; Michon, T.; Walter, J. Electrochemical Atomic Force Microscopy Imaging of Redox-Immunomarked Proteins on Native Potyviruses: From Subparticle to Single-Protein Resolution. ACS Nano 2015, 9, 4911–4924. [Google Scholar] [CrossRef] [PubMed]
- Kueng, A.; Kranz, C.; Lugstein, A.; Bertagnolli, E.; Mizaikoff, B. Integrated AFM-SECM in Tapping Mode: Simultaneous Topographical and Electrochemical Imaging of Enzyme Activity. Angew. Chem. Int. Ed. 2003, 42, 3238–3240. [Google Scholar] [CrossRef] [PubMed]
- Kranz, C.; Kueng, A.; Lugstein, A.; Bertagnolli, E.; Mizaikoff, B. Mapping of enzyme activity by detection of enzymatic products during AFM imaging with integrated SECM-AFM probes. Ultramicroscopy 2004, 100, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Goyer, C.; Anne, A.; Demaille, C. Exploring the Motional Dynamics of End-Grafted DNA Oligonucleotides by in Situ Electrochemical Atomic Force Microscopy. J. Phys. Chem. B 2007, 111, 6051–6058. [Google Scholar] [CrossRef] [PubMed]
- Tavert-Roudet, G.; Anne, A.; Barra, A.; Chovin, A.; Demaille, C.; Michon, T. The potyvirus particle recruits the plant translation initiation factor eIF4E by means of the VPg covalently linked to the viral RNA. Mol. Plant. Microbe Interact. 2017, 30, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Niwa, O.; Maruyama, K.; Shindo, Y.; Oka, K.; Suzuki, K. Neurite Imaging of Living PC12 Cells with Scanning Electrochemical/Near-Field Optical/Atomic Force Microscopy. Angew. Chem. Int. Ed. 2007, 46, 8238–8241. [Google Scholar] [CrossRef] [PubMed]
- Bard, A.J.; Mirkin, M.V.; Unwin, P.R.; Wipf, D.O. Scanning Electrochemical Microscopy. 12. Theory and Experiment of the Feedback Mode with Finite Heterogeneous Electron-Transfer Kinetics and Arbitrary Substrate Size. J. Phys. Chem. 1992, 96, 1861–1868. [Google Scholar] [CrossRef]
- Selzer, Y.; Mandler, D. Scanning Electrochemical Microscopy. Theory of the Feedback Mode for Hemispherical Ultramicroelectrodes: Steady-State and Transient Behavior. Anal. Chem. 2000, 72, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Liljeroth, P.; Johans, C.; Slevin, C.J.; Quinn, B.M.; Kontturi, K. Disk-Generation/Ring-Collection Scanning Electrochemical Microscopy: Theory and Application. Anal. Chem. 2002, 74, 1972–1978. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.D.; Unwin, P.R. Scanning electrochemical microscopy: Theory and experiment for the positive feedback mode with unequal diffusion coefficients of the redox mediator couple. J. Electroanal. Chem. 1997, 439, 123–136. [Google Scholar] [CrossRef]
- Qiu, F.; Fisher, A.C. The boundary element method: Chronoamperometric simulations at microelectrodes. Electrochem. Commun. 2003, 5, 87–93. [Google Scholar] [CrossRef]
- Sklyar, O.; Kueng, A.; Kranz, C.; Mizaikoff, B.; Lugstein, A.; Bertagnolli, E.; Wittstock, G. Numerical Simulation of Scanning Electrochemical Microscopy Experiments with Frame-Shaped Integrated Atomic Force Microscopy−SECM Probes Using the Boundary Element Method. Anal. Chem. 2005, 77, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Pierce, D.T.; Unwin, P.R.; Bard, A.J. Scanning electrochemical microscopy. 17. Studies of enzyme-mediator kinetics for membrane-and surface-immobilized glucose oxidase. Anal. Chem. 1992, 64, 1795–1804. [Google Scholar] [CrossRef]
- Shiku, H.; Takeda, T.; Yamada, H.; Matsue, T.; Uchida, I. Microfabrication and characterization of diaphorase-patterned surfaces by scanning electrochemical microscopy. Anal. Chem. 1995, 67, 312–317. [Google Scholar] [CrossRef]
- Yamada, H.; Fukumoto, H.; Yokoyama, T.; Koike, T. Immobilized Diaphorase Surfaces Observed by Scanning Electrochemical Microscope with Shear Force Based Tip−Substrate Positioning. Anal. Chem. 2005, 77, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Shiku, H.; Hara, Y.; Matsue, T.; Uchida, I.; Yamauchi, T. Dual immunoassay of human chorionic gonadotropin and human placental lactogen at a miocrofabricated substrate by scanning electrochemical microscopy. J. Electroanal. Chem. 1997, 438, 187–190. [Google Scholar] [CrossRef]
- Lei, R.; Stratmann, L.; Schäfer, D.; Erichsen, T.; Neugebauer, S.; Li, N.; Schuhmann, W. Imaging Biocatalytic Activity of Enzyme−Polymer Spots by Means of Combined Scanning Electrochemical Microscopy/Electrogenerated Chemiluminescence. Anal. Chem. 2009, 81, 5070–5074. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wittstock, G. Scanning Electrochemical Microscopy of Quinoprotein Glucose Dehydrogenase. Anal. Chem. 2004, 76, 3145–3154. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, E.; Botz, A.J.R.; Clausmeyer, J.; Wagner, B.; Schuhmann, W.; Stojek, Z.; Nowicka, A.M. Assembling Paramagnetic Ceruloplasmin at Electrode Surfaces Covered with Ferromagnetic Nanoparticles. Scanning Electrochemical Microscopy in the Presence of a Magnetic Field. Langmuir 2015, 31, 8176–8183. [Google Scholar] [CrossRef] [PubMed]
- Gunawan, C.A.; Nam, E.V.; Marquis, C.P.; Gooding, J.J.; Thordarson, P.; Zhao, C. Scanning Electrochemical Microscopy of Cytochrome c Peroxidase through the Orientation-Controlled Immobilisation of Cytochrome c. ChemElectroChem 2016, 3, 1150–1156. [Google Scholar] [CrossRef]
- Eckhard, K.; Schuhmann, W. Alternating current techniques in scanning electrochemical microscopy (AC-SECM). Analyst 2008, 133, 1486–1497. [Google Scholar] [CrossRef] [PubMed]
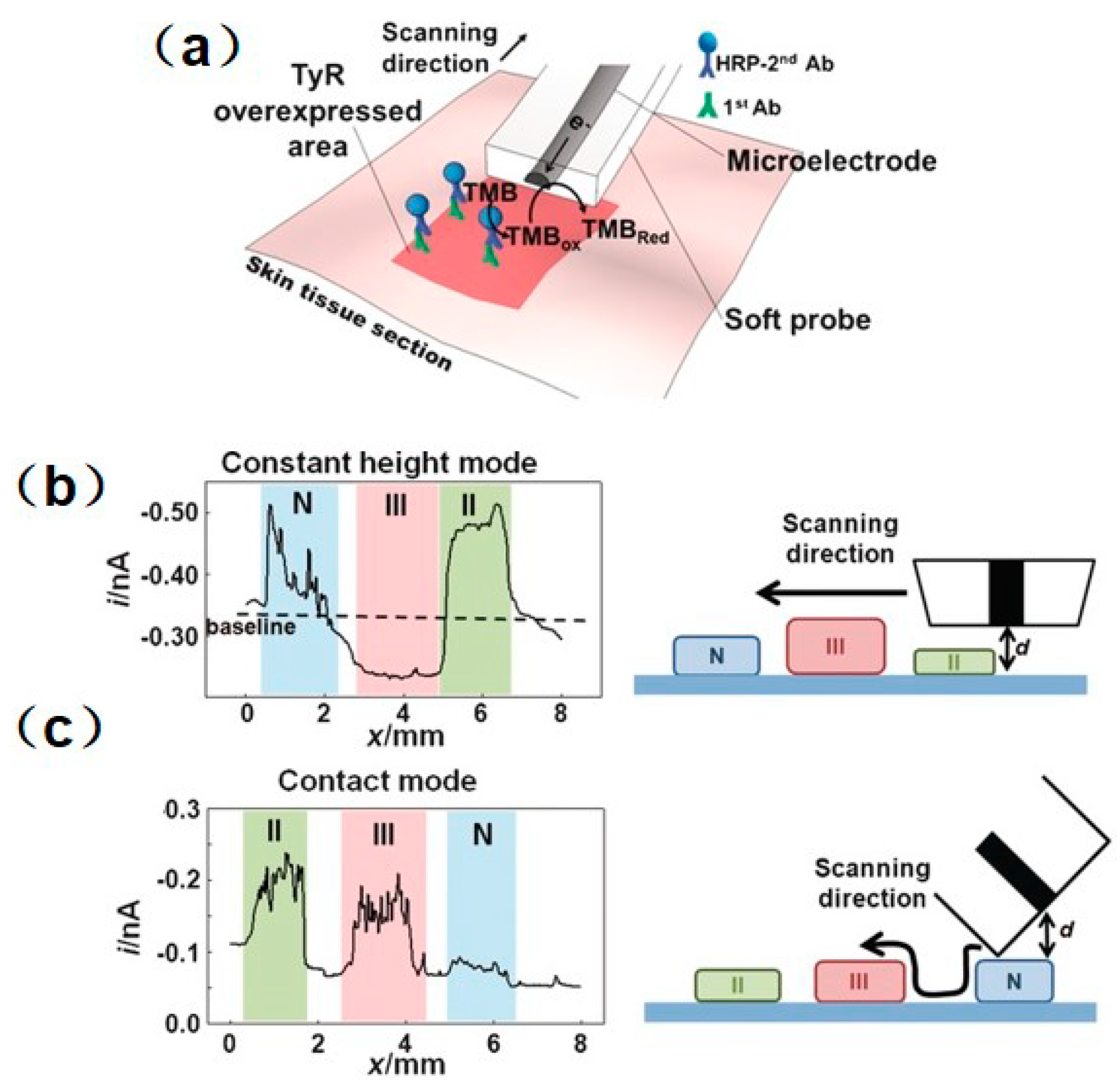
- Lin, T.-E.; Cortés-Salazar, F.; Lesch, A.; Qiao, L.; Bondarenko, A.; Girault, H.H. Multiple scanning electrochemical microscopy mapping of tyrosinase in micro-contact printed fruit samples on polyvinylidene fluoride membrane. Electrochimica Acta 2015, 179, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-E.; Bondarenko, A.; Lesch, A.; Pick, H.; Cortés-Salazar, F.; Girault, H.H. Monitoring Tyrosinase Expression in Non-metastatic and Metastatic Melanoma Tissues by Scanning Electrochemical Microscopy. Angew. Chem. Int. Ed. 2016, 55, 3813–3816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, J.E. Electrochemical detection of single cancer and healthy cell collisions on a microelectrode. Chem. Commun. 2016, 52, 10906–10909. [Google Scholar] [CrossRef] [PubMed]
- Lapenaite, I.; Ramanaviciene, A.; Ramanavicius, A. Current Trends in Enzymatic Determination of Glycerol. Crit. Rev. Anal. Chem. 2006, 36, 13–25. [Google Scholar] [CrossRef]
- Dector, A.; Galindo-de-la-Rosa, J.; Amaya-Cruz, D.M.; Ortíz-Verdín, A.; Guerra-Balcázar, M.; Olivares-Ramírez, J.M.; Arriaga, L.G.; Ledesma-García, J. Towards autonomous lateral flow assays: Paper-based microfluidic fuel cell inside an HIV-test using a blood sample as fuel. Int. J. Hydrog. Energy 2017, 42, 27979–27986. [Google Scholar] [CrossRef]
- Morkvenaite-Vilkonciene, I.; Ramanaviciene, A.; Ramanavicius, A. Redox competition and generation-collection modes based scanning electrochemical microscopy for the evaluation of immobilised glucose oxidase-catalysed reactions. RSC Adv. 2014, 4, 50064–50069. [Google Scholar] [CrossRef]
- Yamashita, K.; Takagi, M.; Takenaka, S.; Uchida, K.; Kondo, H. Visualization of DNA microarrays by scanning electrochemical microscopy (SECM). Analyst 2001, 126, 1210–1211. [Google Scholar] [CrossRef] [PubMed]
- Turcu, F. Imaging immobilised ssDNA and detecting DNA hybridisation by means of the repelling mode of scanning electrochemical microscopy (SECM). Biosens. Bioelectron. 2004, 20, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.S.; Davis, F.; Higson, S.P.J. Scanning electrochemical microscopy of genomic DNA microarrays—Study of adsorption and subsequent interactions. Analyst 2009, 134, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Turcu, F.; Schulte, A.; Hartwich, G.; Schuhmann, W. Label-Free Electrochemical Recognition of DNA Hybridization by Means of Modulation of the Feedback Current in SECM. Angew. Chem. Int. Ed. 2004, 43, 3482–3485. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, M.H.; Kraatz, H.-B. Probing nucleobase mismatch variations by electrochemical techniques: Exploring the effects of position and nature of the single-nucleotide mismatch. Analyst 2010, 135, 2280–2285. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, I.; Laschi, S.; Marrazza, G.; Mascini, M. Electrochemical Imaging of Localized Sandwich DNA Hybridization Using Scanning Electrochemical Microscopy. Anal. Chem. 2007, 79, 7206–7213. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, X.; Jiao, F.; Zhang, F.; Wang, Q.; He, P.; Fang, Y. Scanning Electrochemical Microscopy of DNA Hybridization on DNA Microarrays Enhanced by HRP-Modified SiO2 Nanoparticles. Anal. Chem. 2013, 85, 6511–6517. [Google Scholar] [CrossRef] [PubMed]
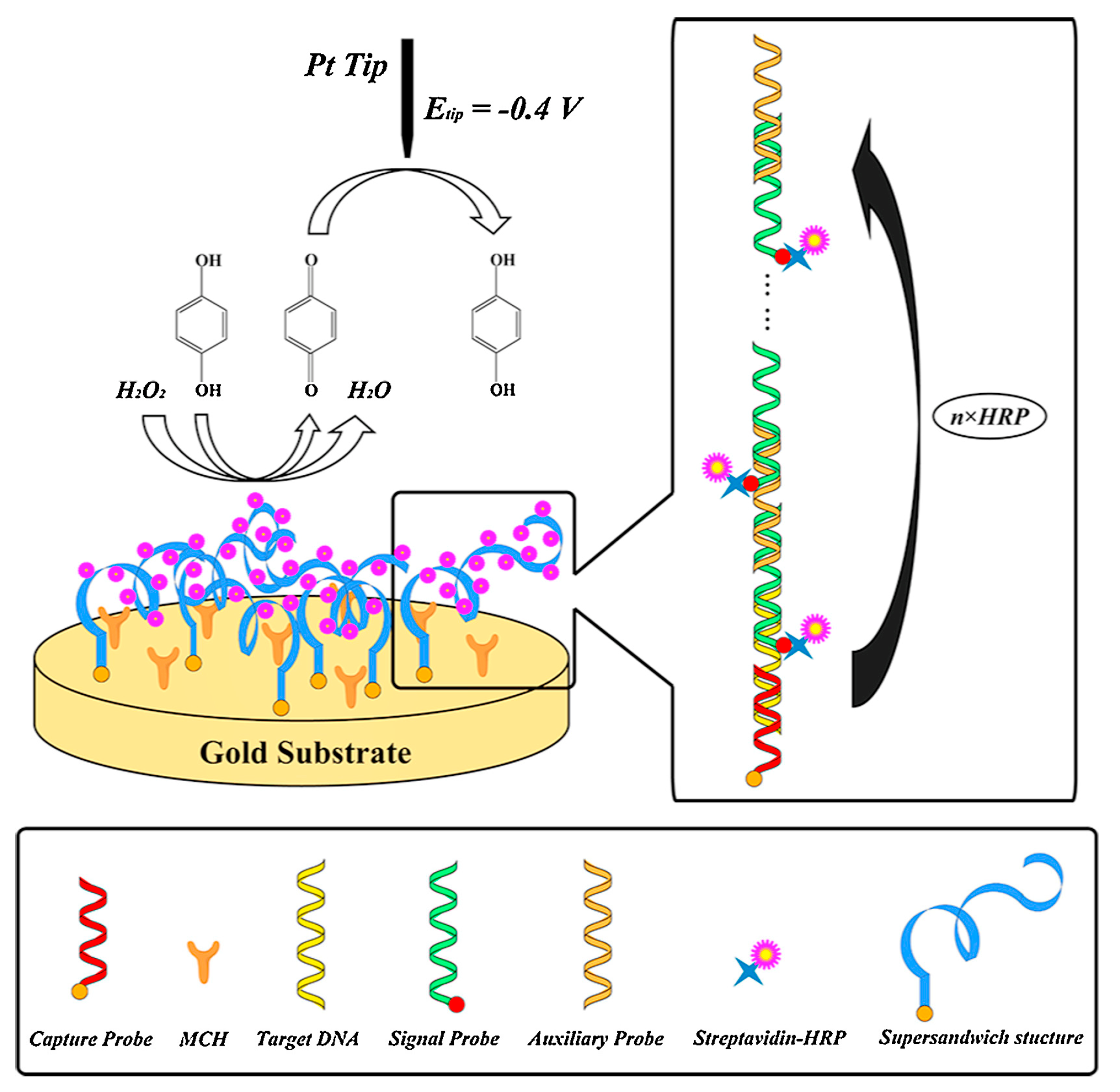
- Chen, B.; Hu, Q.; Xiong, Q.; Zhang, F.; He, P. An ultrasensitive scanning electrochemical microscopy (SECM)-based DNA biosensing platform amplified with the long self-assembled DNA concatemers. Electrochim. Acta 2016, 192, 127–132. [Google Scholar] [CrossRef]
- Rogers, K.R.; Apostol, A.; Madsen, S.J.; Spencer, C.W. Detection of Low Dose Radiation Induced DNA Damage Using Temperature Differential Fluorescence Assay. Anal. Chem. 1999, 71, 4423–4426. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.N.; Schulze, H.; Macdonald, D.; Crain, J.; Mount, A.R.; Bachmann, T.T. Cyclic Denaturation and Renaturation of Double-Stranded DNA by Redox-State Switching of DNA Intercalators. J. Am. Chem. Soc. 2013, 135, 5399–5407. [Google Scholar] [CrossRef] [PubMed]
- Doménech-Carbó, A.; Cebrián-Torrejón, G.; de Miguel, L.; Tordera, V.; Rodrigues-Furtado, D.; Assad-Kahn, S.; Fournet, A.; Figadère, B.; Vázquez-Manrique, R.P.; Poupon, E. dsDNA, ssDNA, G-quadruplex DNA, and nucleosomal DNA electrochemical screening using canthin-6-one alkaloid-modified electrodes. Electrochim. Acta 2014, 115, 546–552. [Google Scholar] [CrossRef]
- Wain, A.J.; Zhou, F. Scanning Electrochemical Microscopy Imaging of DNA Microarrays Using Methylene Blue as a Redox-Active Intercalator. Langmuir 2008, 24, 5155–5160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, J.; Tang, A.; Wu, Z.; Shen, G.; Yu, R. Scanning electrochemical microscopy assay of DNA based on hairpin probe and enzymatic amplification biosensor. Biosens. Bioelectron. 2010, 25, 1953–1957. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.-L.; Mergny, J.-L. Topology of a DNA G-Quadruplex Structure Formed in the HIV-1 Promoter: A Potential Target for Anti-HIV Drug Development. J. Am. Chem. Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.A.; Clark, G.R.; Alexander, S.; Silvestri, G.; Willoughby, C.E. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J. Med. Genet. 2011, 48, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Anne, A.; Cambril, E.; Chovin, A.; Demaille, C. Touching Surface-Attached Molecules with a Microelectrode: Mapping the Distribution of Redox-Labeled Macromolecules by Electrochemical-Atomic Force Microscopy. Anal. Chem. 2010, 82, 6353–6362. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.N.; Shamsi, M.H.; Kraatz, H.-B. Scanning positional variations in single-nucleotide polymorphism of DNA: An electrochemical study. Analyst 2012, 137, 4220–4225. [Google Scholar] [CrossRef] [PubMed]
- Moradi, N.; Noori, A.; Mehrgardi, M.A.; Mousavi, M.F. Scanning Electrochemical Microscopy for Electrochemical Detection of Single-base Mismatches by Tagging Ferrocenecarboxylic Acid as a Redox Probe to DNA. Electroanalysis 2016, 28, 823–832. [Google Scholar] [CrossRef]
- Holmes, J.L.; Davis, F.; Collyer, S.D.; Higson, S.P.J. A new application of scanning electrochemical microscopy for the label-free interrogation of antibody–antigen interactions: Part 2. Anal. Chim. Acta 2012, 741, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.S.; Davis, F.; Holmes, J.L.; Collyer, S.D.; Larcombe, L.D.; Morgan, S.L.; Higson, S.P.J. Detection and imaging the expression of the trans-membrane protein CD44 in RT112 cells by use of enzyme-labeled antibodies and SECM. Biosens. Bioelectron. 2013, 41, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bu, C.; Zhang, Y.; Du, J.; Lu, X.; Liu, X. Composite system based on biomolecules-functionalized multiwalled carbon nanotube and ionic liquid: Electrochemistry and electrocatalysis of tryptophane. Electrochim. Acta 2011, 58, 105–111. [Google Scholar] [CrossRef]
- Polcari, D.; Perry, S.C.; Pollegioni, L.; Geissler, M.; Mauzeroll, J. Localized Detection of d-Serine by using an Enzymatic Amperometric Biosensor and Scanning Electrochemical Microscopy. ChemElectroChem 2017, 4, 920–926. [Google Scholar] [CrossRef]
- Alizadeh, V.; Mousavi, M.F.; Mehrgardi, M.A.; Kazemi, S.H.; Sharghi, H. Electron transfer kinetics of cytochrome c immobilized on a phenolic terminated thiol self assembled monolayer determined by scanning electrochemical microscopy. Electrochim. Acta 2011, 56, 6224–6229. [Google Scholar] [CrossRef]
- Alizadeh, V.; Mehrgardi, M.A.; Fazlollah Mousavi, M. Electrochemical Investigation of Cytochrome c Immobilized onto Self-Assembled Monolayer of Captopril. Electroanalysis 2013, 25, 1689–1696. [Google Scholar] [CrossRef]
- Song, W.; Yan, Z.; Hu, K. Electrochemical immunoassay for CD10 antigen using scanning electrochemical microscopy. Biosens. Bioelectron. 2012, 38, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Kasai, S.; Yokota, A.; Zhou, H.; Nishizawa, M.; Niwa, K.; Onouchi, T.; Matsue, T. Immunoassay of the MRSA-Related Toxic Protein, Leukocidin, with Scanning Electrochemical Microscopy. Anal. Chem. 2000, 72, 5761–5765. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, M.E.; Piantavigna, S.; Bond, A.M.; Graham, B.; Spiccia, L.; Martin, L.L.; O’Mullane, A.P. An SECM study on the influence of cationic, membrane-active peptides on a gold-supported self-assembled monolayer. Electrochem. Commun. 2015, 51, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Piantavigna, S.; Abdelhamid, M.E.; Zhao, C.; Qu, X.; McCubbin, G.A.; Graham, B.; Spiccia, L.; O’Mullane, A.P.; Martin, L.L. Mechanistic Details of the Membrane Perforation and Passive Translocation of TAT Peptides. ChemPlusChem 2015, 80, 83–90. [Google Scholar] [CrossRef]
- She, Z.; Topping, K.; Shamsi, M.H.; Wang, N.; Chan, N.W.C.; Kraatz, H.-B. Investigation of the Utility of Complementary Electrochemical Detection Techniques to Examine the in Vitro Affinity of Bacterial Flagellins for a Toll-Like Receptor 5 Biosensor. Anal. Chem. 2015, 87, 4218–4224. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Hirano, Y.; Motochi, N.; Shiku, H.; Matsue, T. Enzyme immunosensing of pepsinogens 1 and 2 by scanning electrochemical microscopy. Biosens. Bioelectron. 2007, 22, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Xiong, Q.; Zhang, F.; He, P. Simultaneous detection of tumor markers in lung cancer using scanning electrochemical microscopy. J. Electroanal. Chem. 2018, 812, 101–106. [Google Scholar] [CrossRef]
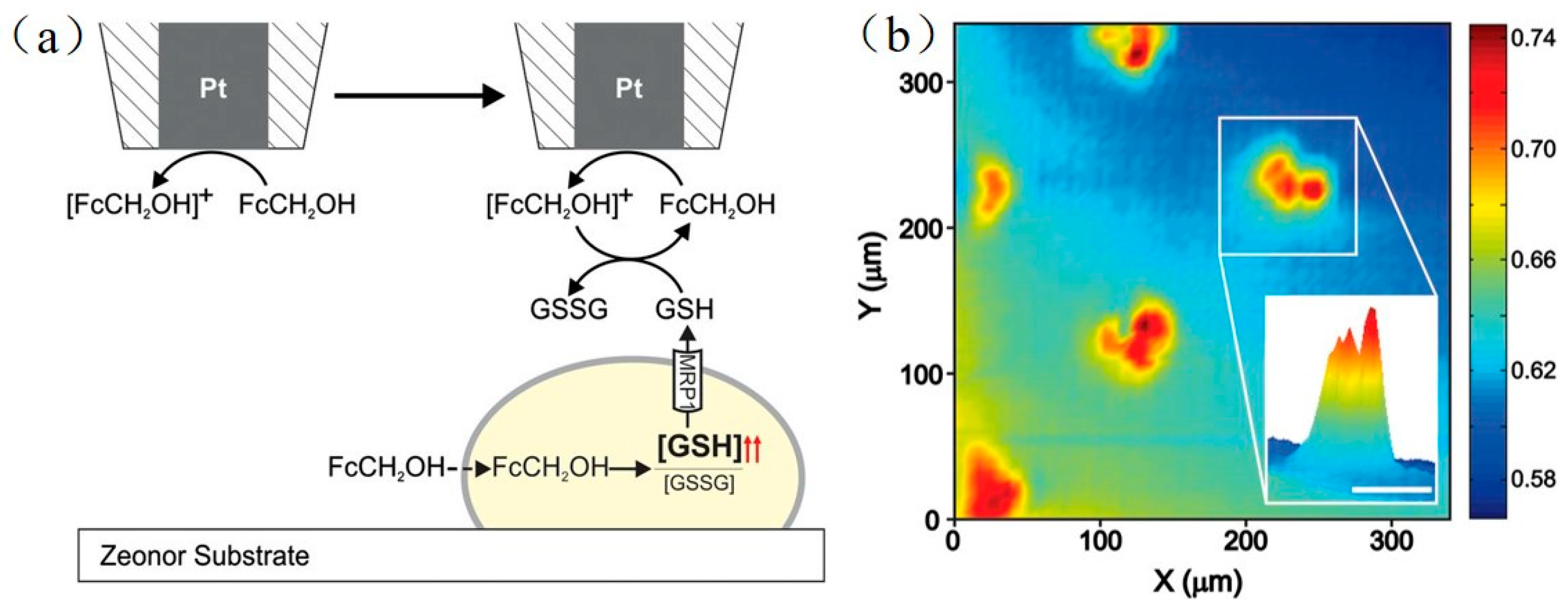
- Kuss, S.; Polcari, D.; Geissler, M.; Brassard, D.; Mauzeroll, J. Assessment of multidrug resistance on cell coculture patterns using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 9249–9254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, J.L.; Davis, F.; Collyer, S.D.; Higson, S.P.J. A new application of scanning electrochemical microscopy for the label-free interrogation of antibody–antigen interactions. Anal. Chim. Acta 2011, 689, 206–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souto, D.E.P.; Silva, J.V.; Martins, H.R.; Reis, A.B.; Luz, R.C.S.; Kubota, L.T.; Damos, F.S. Development of a label-free immunosensor based on surface plasmon resonance technique for the detection of anti-Leishmania infantum antibodies in canine serum. Biosens. Bioelectron. 2013, 46, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Armbrecht, L.; Dittrich, P.S. Recent Advances in the Analysis of Single Cells. Anal. Chem. 2017, 89, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Rotenberg, S.A.; Mirkin, M.V. Scanning electrochemical microscopy of living cells: Different redox activities of nonmetastatic and metastatic human breast cells. Proc. Natl. Acad. Sci. USA 2000, 97, 9855–9860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, M.; Takoh, K.; Matsue, T. Micropatterning of HeLa cells on glass substrates and evaluation of respiratory activity using microelectrodes. Langmuir 2002, 18, 3645–3649. [Google Scholar] [CrossRef]
- Zhao, X.; Lam, S.; Jass, J.; Ding, Z. Scanning electrochemical microscopy of single human urinary bladder cells using reactive oxygen species as probe of inflammatory response. Electrochem. Commun. 2010, 12, 773–776. [Google Scholar] [CrossRef]
- Kikuchi, H.; Prasad, A.; Matsuoka, R.; Aoyagi, S.; Matsue, T.; Kasai, S. Scanning Electrochemical Microscopy Imaging during Respiratory Burst in Human Cell. Front. Physiol. 2016, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Ktari, N.; Poncet, P.; Sénéchal, H.; Malaquin, L.; Kanoufi, F.; Combellas, C. Patterning of Polystyrene by Scanning Electrochemical Microscopy. Biological Applications to Cell Adhesion. Langmuir 2010, 26, 17348–17356. [Google Scholar] [CrossRef] [PubMed]
- Bergner, S.; Palatzky, P.; Wegener, J.; Matysik, F.-M. High-Resolution Imaging of Nanostructured Si/SiO2 Substrates and Cell Monolayers Using Scanning Electrochemical Microscopy. Electroanalysis 2011, 23, 196–200. [Google Scholar] [CrossRef]
- Shiku, H.; Shiraishi, T.; Aoyagi, S.; Utsumi, Y.; Matsudaira, M.; Abe, H.; Hoshi, H.; Kasai, S.; Ohya, H.; Matsue, T. Respiration activity of single bovine embryos entrapped in a cone-shaped microwell monitored by scanning electrochemical microscopy. Anal. Chim. Acta 2004, 522, 51–58. [Google Scholar] [CrossRef]
- Barker, A.L.; Macpherson, J.V.; Slevin, C.J.; Unwin, P.R. Scanning Electrochemical Microscopy (SECM) as a Probe of Transfer Processes in Two-Phase Systems: Theory and Experimental Applications of SECM-Induced Transfer with Arbitrary Partition Coefficients, Diffusion Coefficients, and Interfacial Kinetics. J. Phys. Chem. B 1998, 102, 1586–1598. [Google Scholar] [CrossRef]
- Utsunomiya, T.; Goto, K.; Nasu, M.; Kumasako, Y.; Araki, Y.; Yokoo, M.; Itohsasaki, T.; Abe, H. Evaluating the Quality of Human Embryos with a Measurement of Oxygen Consumption by Scanning Electrochemical Microscopy. J. Mamm. Ova Res. 2016, 25, 2–7. [Google Scholar] [CrossRef]
- Matsumae, Y.; Takahashi, Y.; Ino, K.; Shiku, H.; Matsue, T. Electrochemical monitoring of intracellular enzyme activity of single living mammalian cells by using a double-mediator system. Anal. Chim. Acta 2014, 842, 20–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
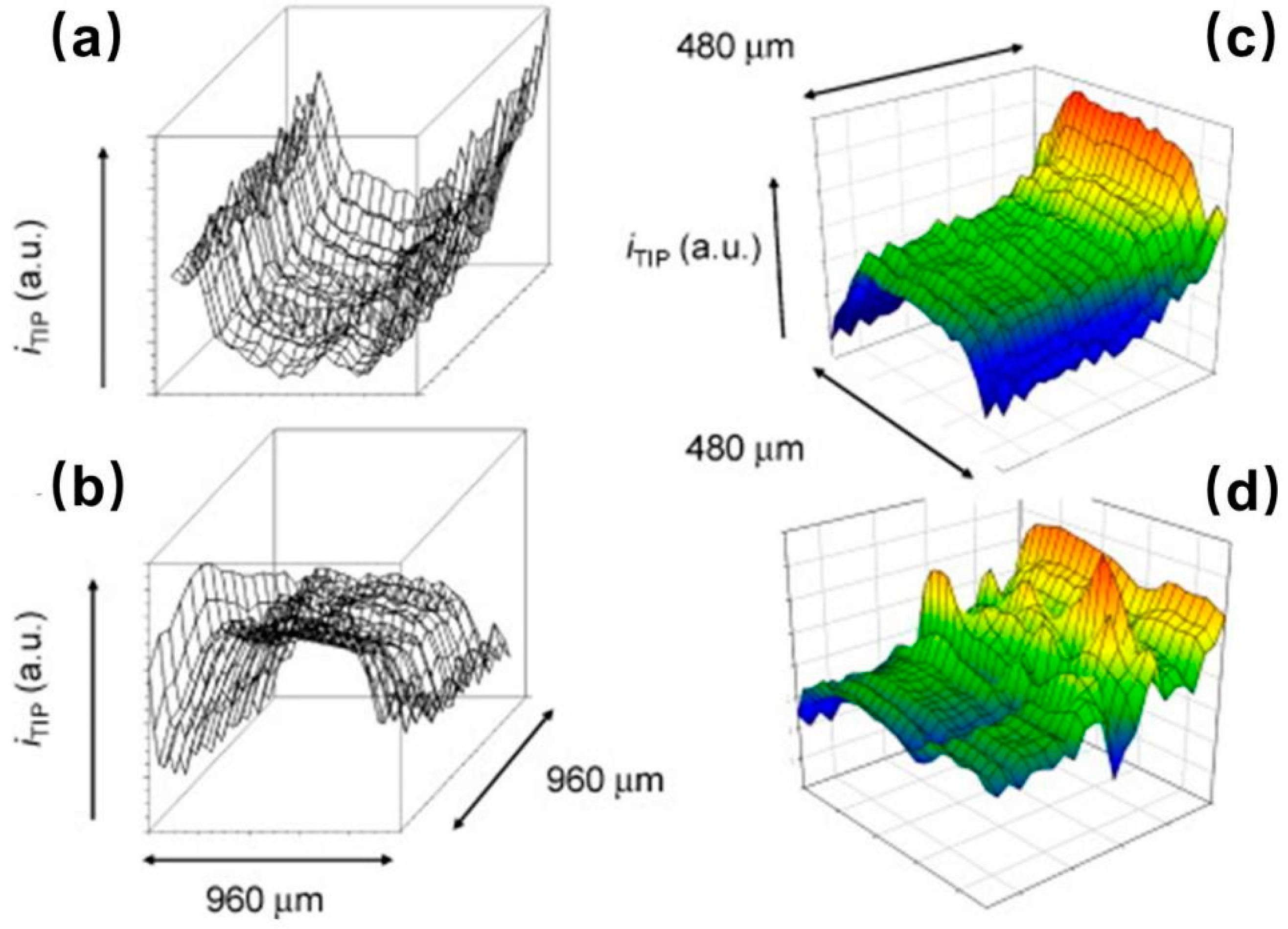
- Zhao, X.; Diakowski, P.M.; Ding, Z. Deconvoluting Topography and Spatial Physiological Activity of Live Macrophage Cells by Scanning Electrochemical Microscopy in Constant-Distance Mode. Anal. Chem. 2010, 82, 8371–8373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.-N.; Long, Y.-T.; Ding, Z. Filming a live cell by scanning electrochemical microscopy: Label-free imaging of the dynamic morphology in real time. Chem. Cent. J. 2012, 6, 20. [Google Scholar] [CrossRef] [PubMed]
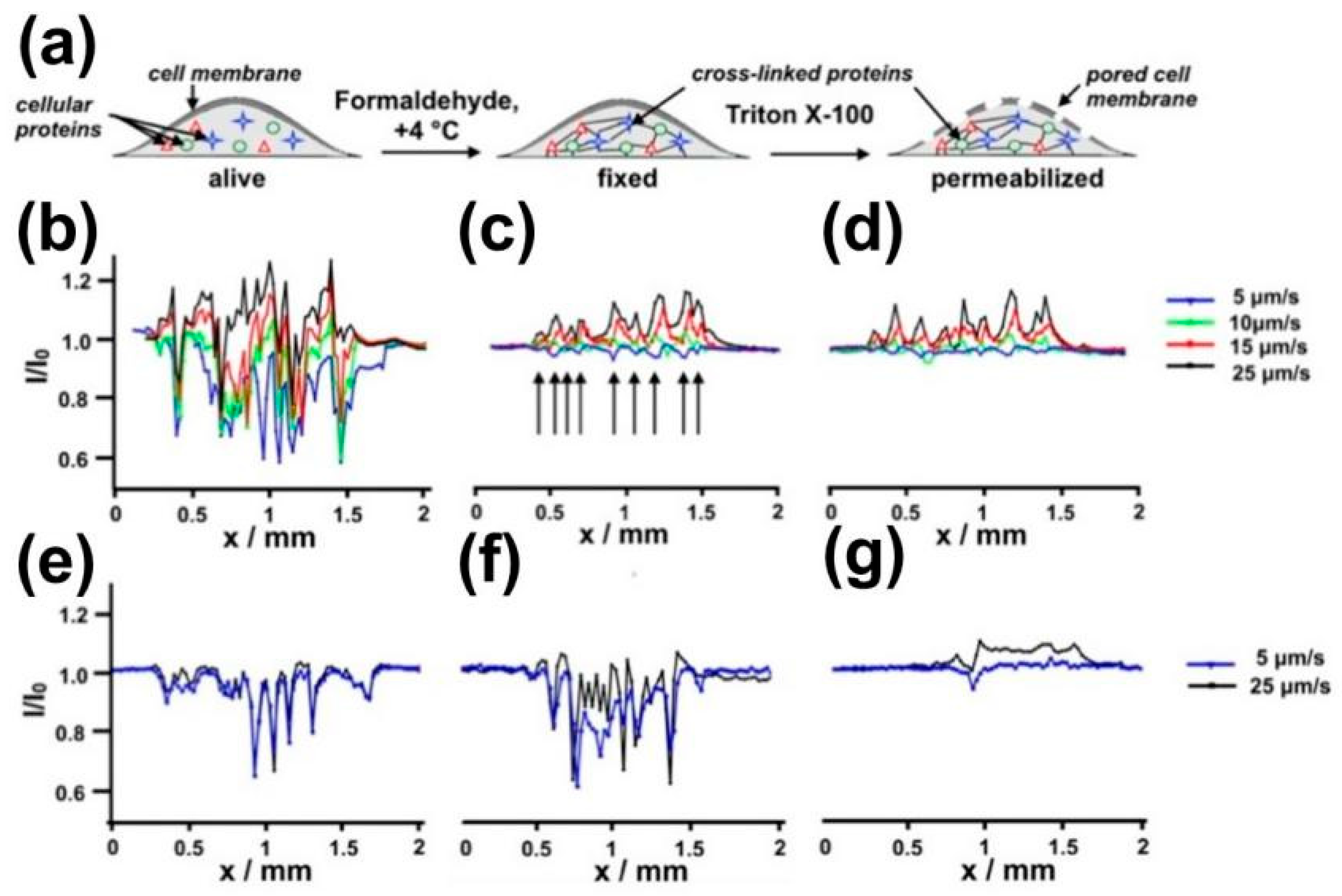
- Bondarenko, A.; Lin, T.E.; Stupar, P.; Lesch, A.; Cortéssalazar, F.; Girault, H.H.; Pick, H. Fixation and Permeabilization Approaches for Scanning Electrochemical Microscopy of Living Cells. Anal. Chem. 2016, 88, 11436–11443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
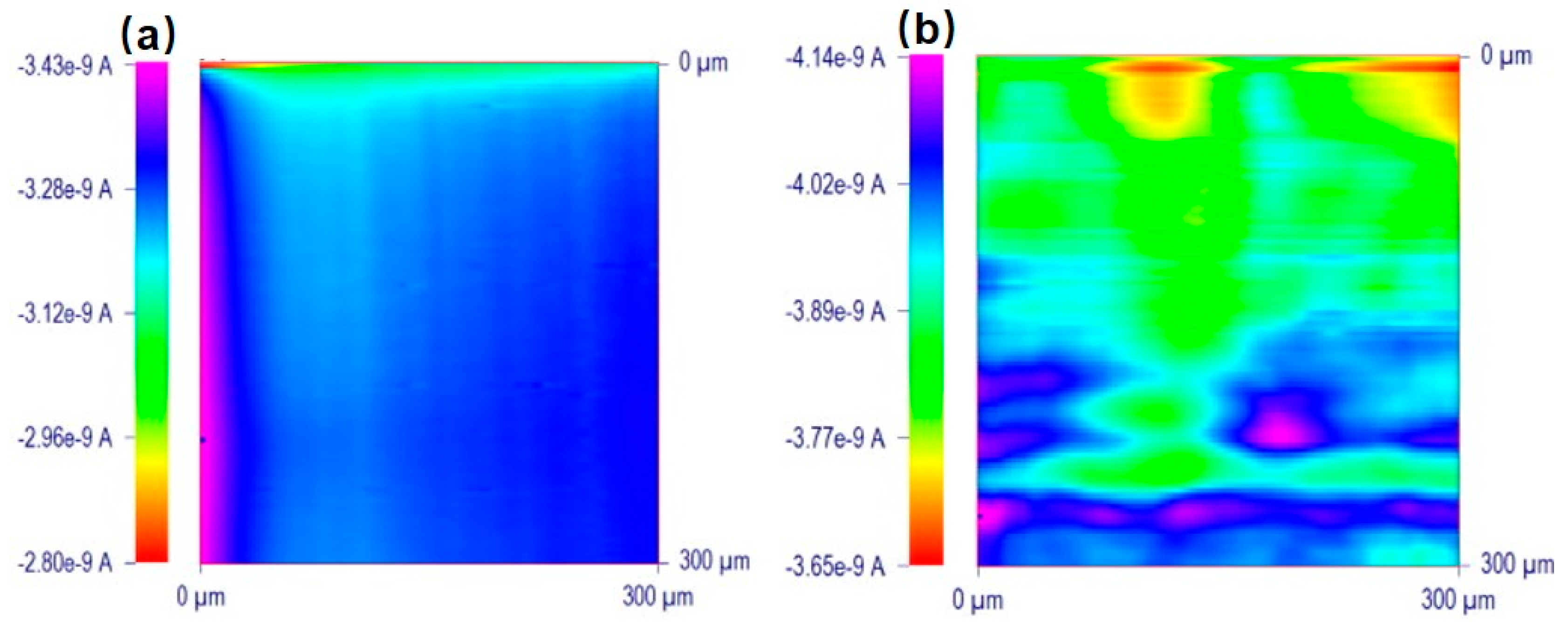
- Li, M.S.M.; Filice, F.P.; Henderson, J.D.; Ding, Z. Probing Cd2+-Stressed Live Cell Membrane Permeability with Various Redox Mediators in Scanning Electrochemical Microscopy. J. Phys. Chem. C 2016, 120, 6094–6103. [Google Scholar] [CrossRef]
- Henderson, J.D.; Filice, F.P.; Li, M.S.M.; Ding, Z. Tracking live cell response to cadmium (II) concentrations by scanning electrochemical microscopy. J. Inorg. Biochem. 2016, 158, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Filice, F.P.; Li, M.S.M.; Henderson, J.D.; Ding, Z. Mapping Cd2+-induced membrane permeability changes of single live cells by means of scanning electrochemical microscopy. Anal. Chim. Acta 2016, 908, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Abrahamson, M.; Lewandowski, Z.; Geesey, G.; Skjak-Braek, G.; Strand, W.; Christensen, B.E. Development of an artificial biofilm to study the effects of a single microcolony on mass transport. J. Microbiol. Methods 1996, 26, 161–169. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Wang, J. Heterogeneous electrochemical characteristics of biofilm/metal interface and local electrochemical techniques used for this purpose. Mater. Corros. 2009, 60, 957–962. [Google Scholar] [CrossRef]
- Tan, S.Y.-E.; Chew, S.C.; Tan, S.Y.-Y.; Givskov, M.; Yang, L. Emerging frontiers in detection and control of bacterial biofilms. Curr. Opin. Biotechnol. 2014, 26, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wessel, A.K.; Hmelo, L.; Parsek, M.R.; Whiteley, M. Going local: Technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 2013, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- And, K.B.H.; Bard, A.J. Interaction of Silver(I) Ions with the Respiratory Chain of Escherichia coli: An Electrochemical and Scanning Electrochemical Microscopy Study of the Antimicrobial Mechanism of Micromolar Ag+. Biochemistry 2005, 44, 13214–13223. [Google Scholar]
- Nagamine, K.; Onodera, S.; Kurihara, A.; Yasukawa, T.; Shiku, H.; Asano, R.; Kumagai, I.; Matsue, T. Electrochemical screening of recombinant protein solubility inEscherichia coli using scanning electrochemical microscopy (SECM). Biotechnol. Bioeng. 2007, 96, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Kaya, T.; Yasukawa, T.; Shiku, H.; Matsue, T. Application of microbial chip for amperometric detection of metabolic alteration in bacteria. Sens. Actuators B Chem. 2005, 108, 676–682. [Google Scholar] [CrossRef]
- Matsui, N.; Kaya, T.; Nagamine, K.; Yasukawa, T.; Shiku, H.; Matsue, T. Electrochemical mutagen screening using microbial chip. Biosens. Bioelectron. 2006, 21, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Liu, B.; Mirkin, M.V.; Frank, H.A.; Rusling, J.F. Scanning electrochemical microscopy of living cells. 3. Rhodobacter s phaeroides. Anal. Chem. 2002, 74, 114–119. [Google Scholar] [PubMed]
- Zhang, W.; Wu, H.; Hsing, I.-M. Real-Time Label-Free Monitoring of Shewanella oneidensis MR-1 Biofilm Formation on Electrode During Bacterial Electrogenesis Using Scanning Electrochemical Microscopy. Electroanalysis 2015, 27, 648–655. [Google Scholar] [CrossRef]
- Harris, D.; Ummadi, J.G.; Thurber, A.R.; Allau, Y.; Verba, C.; Colwell, F.; Torres, M.E.; Koley, D. Real-time monitoring of calcification process by Sporosarcina pasteurii biofilm. Analyst 2016, 141, 2887–2895. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
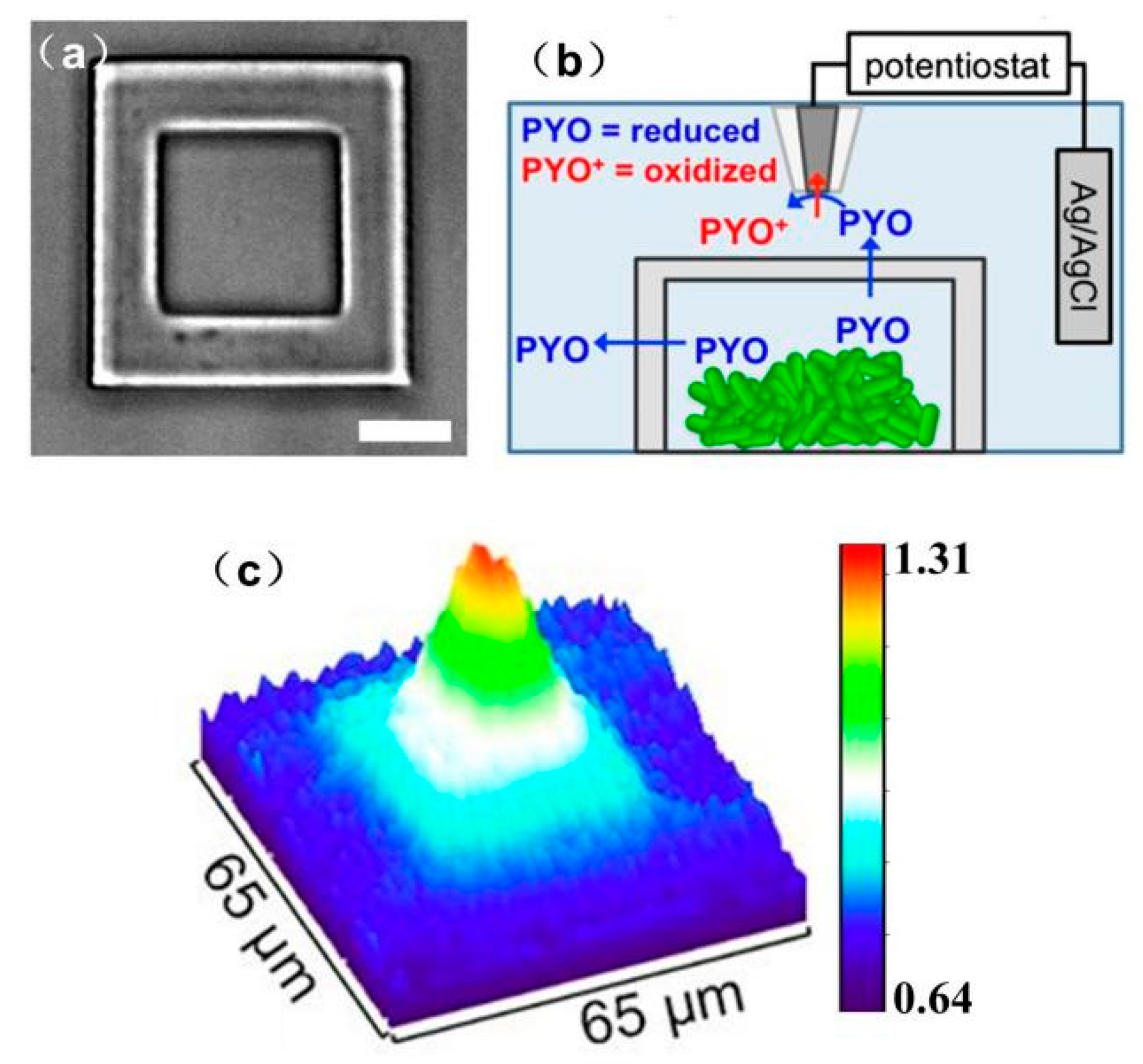
- Connell, J.; Kim, J.; Shear, J.B.; Bard, A.J.; Whiteley, M. Analyzing Secondary Metabolite Production by 3D Printed Bacterial Populations Using Scanning Electrochemical Microscopy. Microsc. Microanal. 2014, 20, 1182–1183. [Google Scholar] [CrossRef]
- Connell, J.L.; Kim, J.; Shear, J.B.; Bard, A.J.; Whiteley, M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 18255–18260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
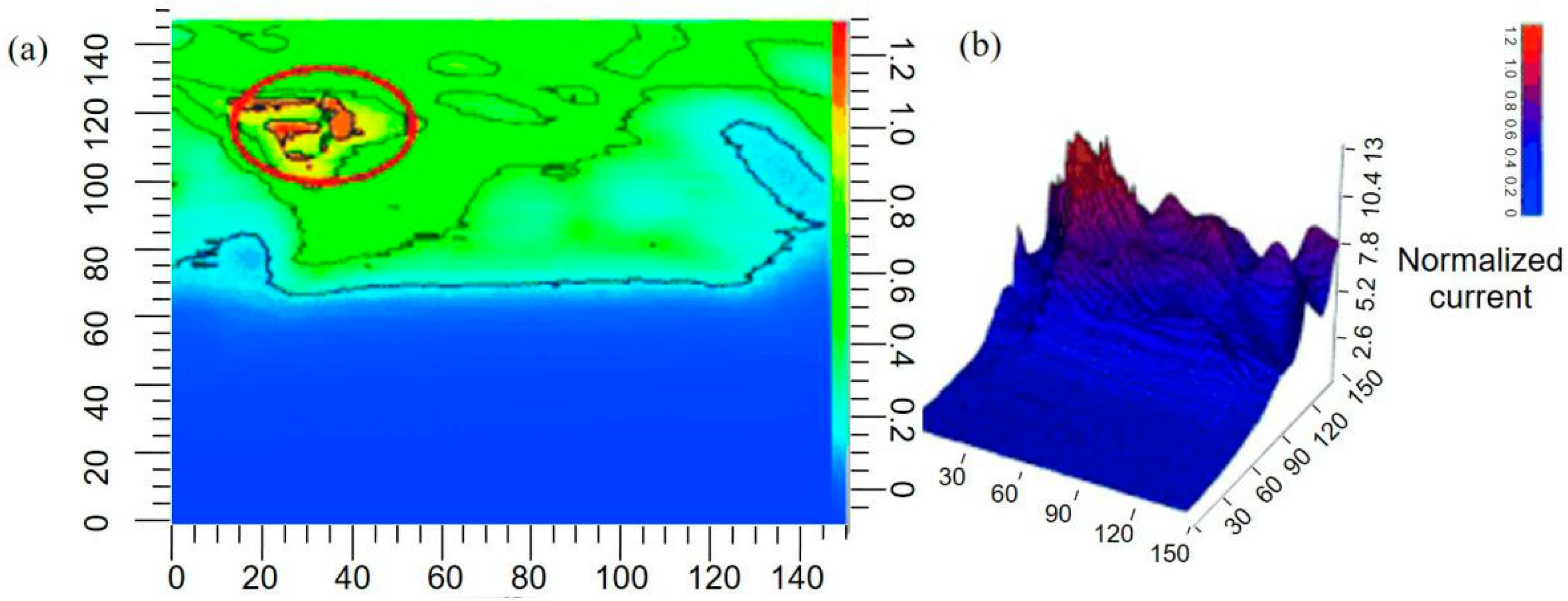
- Liu, X.; Ramsey, M.M.; Chen, X.; Koley, D.; Whiteley, M.; Bard, A.J. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 2668–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
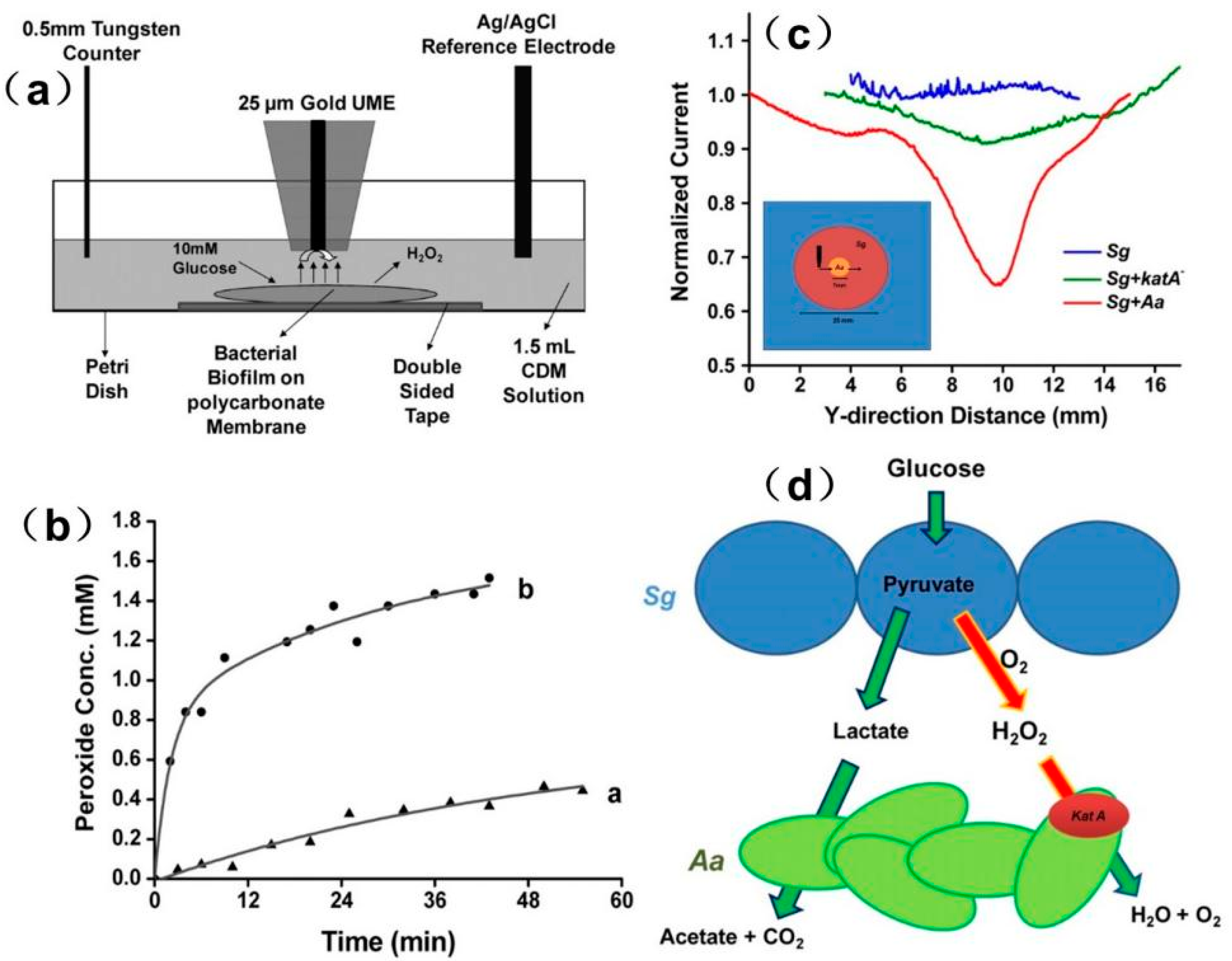
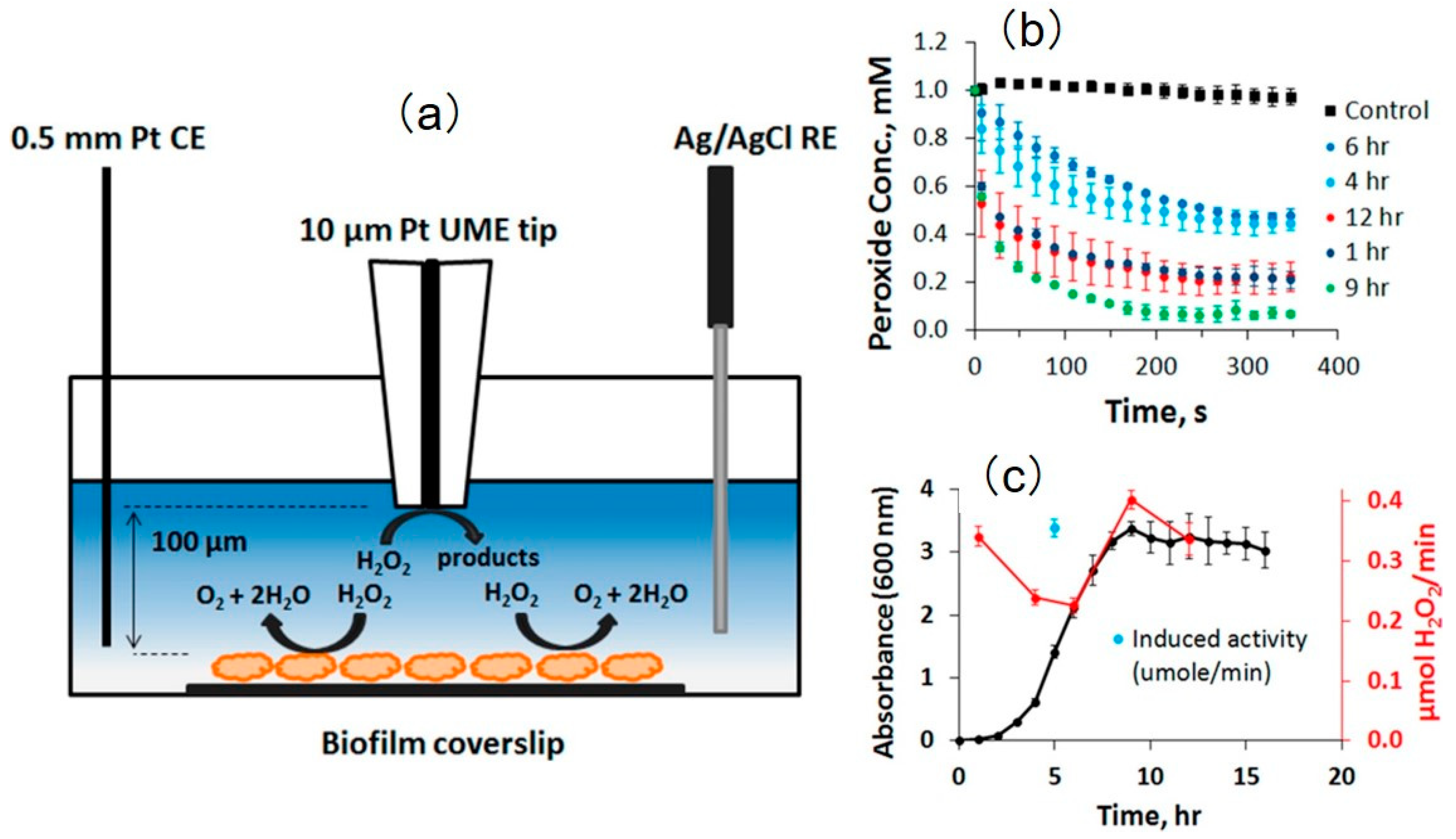
- Abucayon, E.; Ke, N.; Cornut, R.; Patelunas, A.; Miller, D.; Nishiguchi, M.K.; Zoski, C.G. Investigating Catalase Activity Through Hydrogen Peroxide Decomposition by Bacteria Biofilms in Real Time Using Scanning Electrochemical Microscopy. Anal. Chem. 2014, 86, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Videla, H.A. Manual of Biocorrosion; CRC Press: Boca Raton, FL, USA, 1996; ISBN 978-0-87371-726-7. [Google Scholar]
- Venzlaff, H.; Enning, D.; Srinivasan, J.; Mayrhofer, K.J.J.; Hassel, A.W.; Widdel, F.; Stratmann, M. Accelerated cathodic reaction in microbial corrosion of iron due to direct electron uptake by sulfate-reducing bacteria. Corros. Sci. 2013, 66, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Zhu, Z.; Chang, L.; Zhang, J.; Cao, F.; Zhang, J. An in-situ Degradation Behavior Study of MAO Coating on AZ91D Magnesium Alloy in Aqueous Solutions by SECM. Int. J. Electrochem. Sci. 2017, 2145–2158. [Google Scholar] [CrossRef]
- Qian, H.; Li, M.; Li, Z.; Lou, Y.; Huang, L.; Zhang, D.; Xu, D.; Du, C.; Lu, L.; Gao, J. Mussel-inspired superhydrophobic surfaces with enhanced corrosion resistance and dual-action antibacterial properties. Mater. Sci. Eng. C 2017, 80, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.; Schütz, M.K.; Libert, M.; Tribollet, B.; Vivier, V. Influence of hydrogen-oxidizing bacteria on the corrosion of low carbon steel: Local electrochemical investigations. Bioelectrochemistry 2014, 97, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busalmen, J.P.; Vazquez, M.; De Sanchez, S.R. New evidences on the catalase mechanism of microbial corrosion. Electrochim. Acta 2002, 47, 1857–1865. [Google Scholar] [CrossRef]
- Baeza, S.; Vejar, N.; Gulppi, M.; Azocar, M.; Melo, F.; Monsalve, A.; Pérez-Donoso, J.; Vásquez, C.C.; Pavez, J.; Zagal, J.H.; et al. New evidence on the role of catalase in Escherichia coli-mediated biocorrosion. Corros. Sci. 2013, 67, 32–41. [Google Scholar] [CrossRef]
- Momotenko, D.; Byers, J.C.; McKelvey, K.; Kang, M.; Unwin, P.R. High-Speed Electrochemical Imaging. ACS Nano 2015, 9, 8942–8952. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.; Trinh, D.; Mauzeroll, J. High-Speed Scanning Electrochemical Microscopy Method for Substrate Kinetic Determination: Application to Live Cell Imaging in Human Cancer. Anal. Chem. 2015, 87, 8102–8106. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Li, Z.; Lou, Y.; Cao, F.; Zhang, D.; Li, X. Recent Advances in Scanning Electrochemical Microscopy for Biological Applications. Materials 2018, 11, 1389. https://doi.org/10.3390/ma11081389
Huang L, Li Z, Lou Y, Cao F, Zhang D, Li X. Recent Advances in Scanning Electrochemical Microscopy for Biological Applications. Materials. 2018; 11(8):1389. https://doi.org/10.3390/ma11081389
Chicago/Turabian StyleHuang, Luyao, Ziyu Li, Yuntian Lou, Fahe Cao, Dawei Zhang, and Xiaogang Li. 2018. "Recent Advances in Scanning Electrochemical Microscopy for Biological Applications" Materials 11, no. 8: 1389. https://doi.org/10.3390/ma11081389



