Effects of Oenanthe javanica on Nitrogen Removal in Free-Water Surface Constructed Wetlands under Low-Temperature Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Water and Plant Sampling and Analysis
2.3. Microorganism Sampling and Analysis
2.3.1. Preparation of Microbial Samples
2.3.2. Extraction of Total Genomic DNA
2.3.3. Real-Time Quantitative PCR Analysis
2.3.4. 16.S rRNA Gene Illumina MiSeq Sequencing
2.3.5. Sequence Storage Information
2.4. Statistical Analysis
3. Results
3.1. Nutrient Removal Performance
3.2. Plant Growth Dynamics and Physiological Root Characteristics
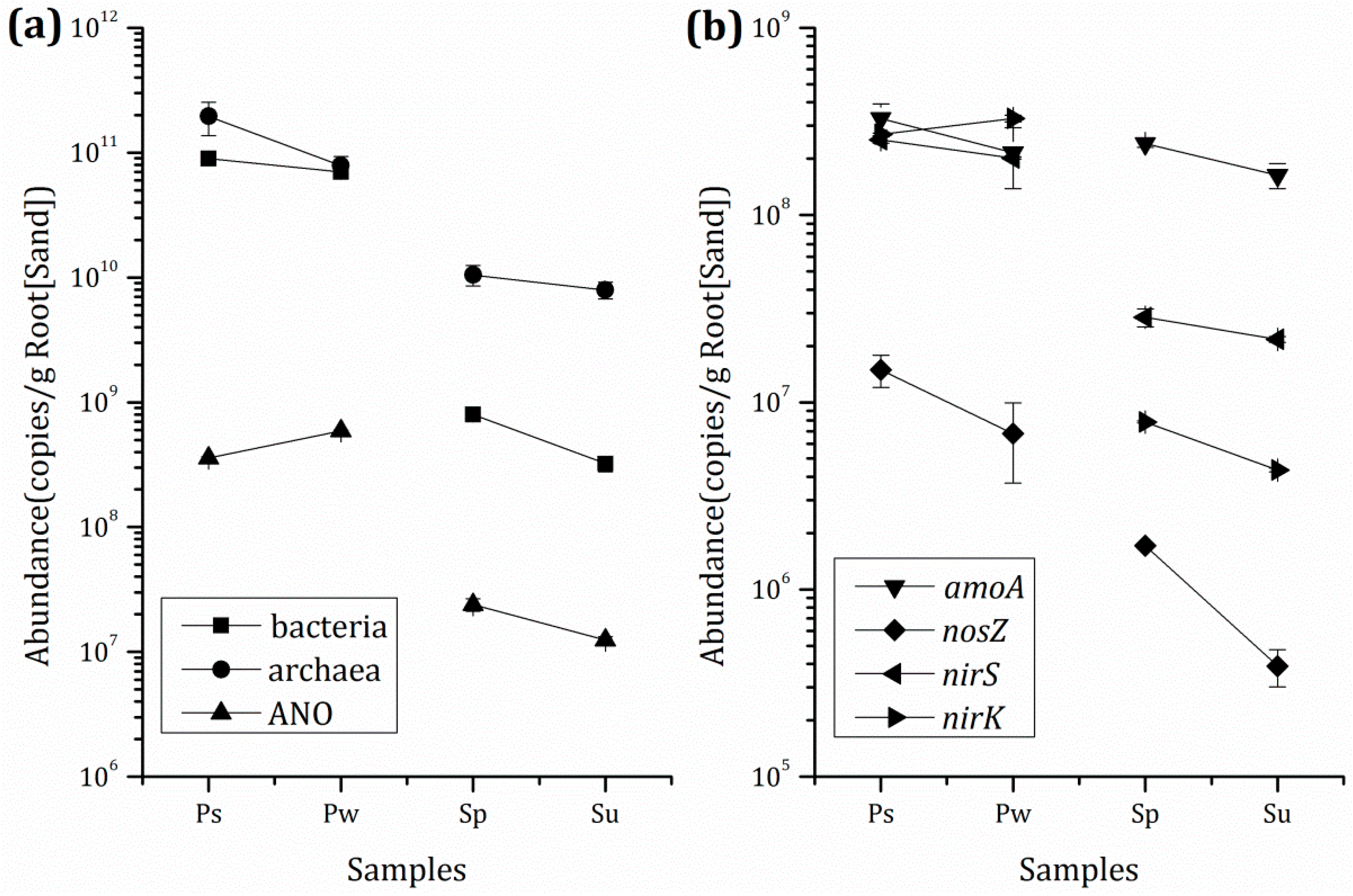
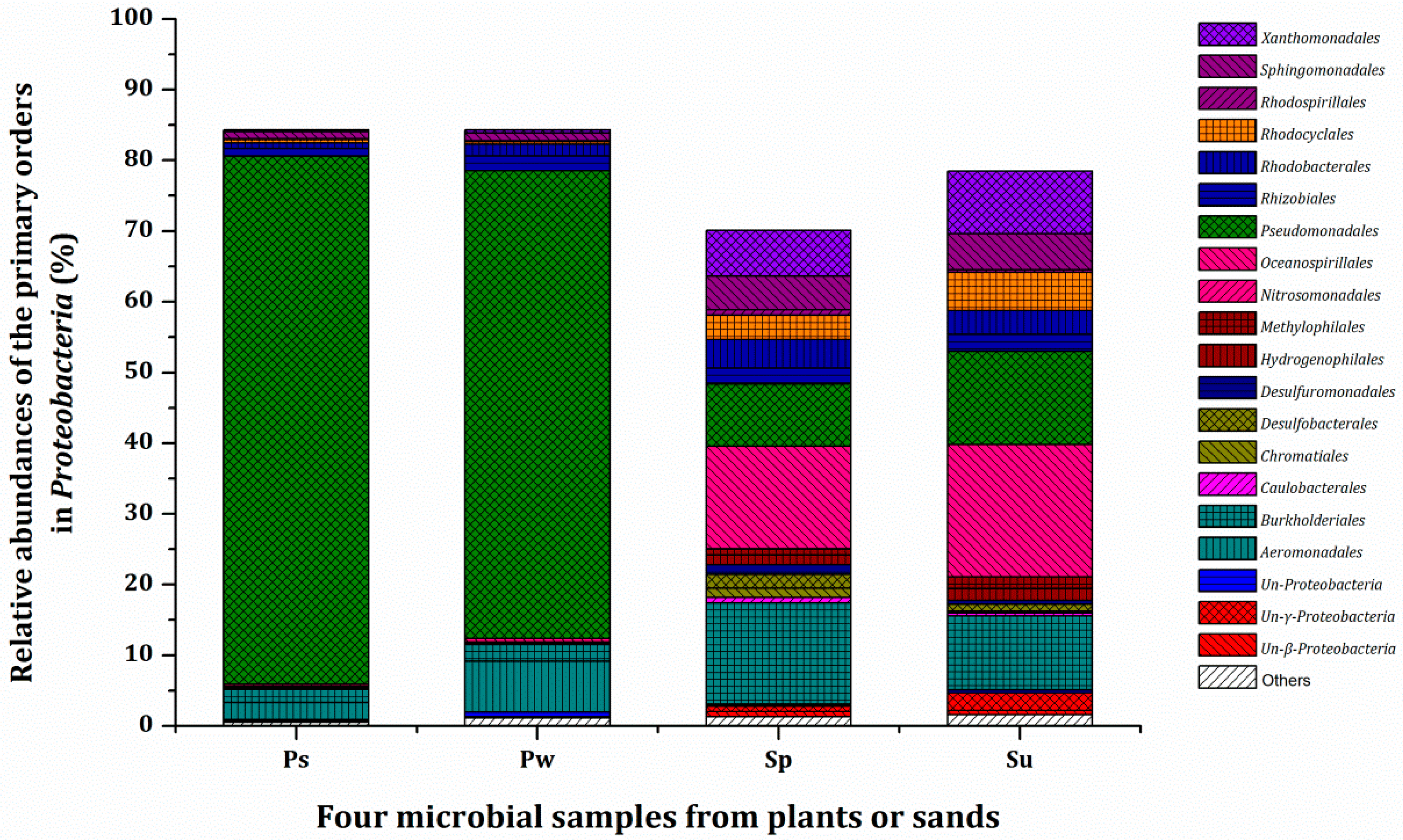
3.3. Microbial Population and Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Target | Primer | Primer Sequence (5′-3′) | References |
|---|---|---|---|
| bacterial 16S rRNA gene | 690F | TGTGTAGCGGTGAAATGCG | [65] |
| 829R | CATCGTTTACGGCGTGGAC | ||
| archaeal 16S rRNA gene | ARC344F | ACGGGGYGCAGCAGGCGCGA | [66] |
| ARC915R | GTGCTCCCCCGCCAATTCCT | ||
| ANO 16S rRNA gene | AMX809F | GCCGTAAACGATGGGCACT | [67] |
| AMX1066R | AACGTCTCACGACACGAGCTG | ||
| amoA | amo1F | GGGGGTTTCTACTGGTGGT | [68] |
| amo2R | CCCCTCKGSAAAGCCTTCTTC | ||
| nosZ | NosZ 1527F | CGCTGTTCHTCGACAGYCA | [69] |
| NosZ 1773R | ATRTCGATCARCTGBTCGTT | ||
| nirS | nirS cd3AF | GTSAACGTSAAGGARACSGG | [70] |
| nirS R3cd | GASTTCGGRTGSGTCTTGA | ||
| nirK | nirK 583F | TCATGGTGCTGCCGCGKGACGG | [71] |
| nirK 909R | GAACTTGCCGGTKGCCCAGAC |
| Information | qPCR | Illumina MiSeq Sequencing |
|---|---|---|
| Analysis system | Illumina-Eco real-time PCR system (Illumina, San Diego, CA, USA) | Illumina MiSeq 2500 sequencing platform (Illumina, San Diego, CA, USA) |
| Reaction mixture | 5.0 μL SYBR® Premix Ex Taq™ II (Takara, Otsu, Japan), 1.0 μL template DNA (diluted 100-fold), 0.5 μL forward and 0.5 μL reverse primers (10 μM), 3.0 μL RNase-free water | 25 μL reaction mixture (including 10 ng template, 0.5 μL forward primer, 0.5 μL reverse primer) |
| PCR program | 30 s at 94 °C, 40 cycles of 5 s at 95 °C, 30 s at 55 °C (amoA) or 60 °C (other genes), and 30 s at 72 °C | 3 min at 94 °C, 30 cycles of 10 s at 94 °C, 15 s at 55 °C, and 72 °C for 30 s, and a final incubation at 72 °C for 7 min |
| PCR product purification | / | Agencourt AMPure beads (Beckman Coulter, Inc., Fullerton, CA, USA) |
| Libraries construction | / | NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs Inc., Boston, MA, USA) |
| Sample | OTUs | ACE | Simpson | Shannon-Even | Coverage |
|---|---|---|---|---|---|
| Ps | 446 | 629.398681 | 0.219313 | 0.382270 | 0.994148 |
| Pw | 567 | 720.449124 | 0.137883 | 0.462411 | 0.994037 |
| Sp | 710 | 788.302182 | 0.031945 | 0.748674 | 0.995926 |
| Su | 675 | 761.131090 | 0.046425 | 0.695539 | 0.995444 |
| Phylum | Genus | Read Numbers | Relative Abundances (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ps | Pw | Sp | Su | Ps | Pw | Sp | Su | ||
| Bacteroidetes | Anaerorhabdus | 0 | 5 | 278 | 98 | 0.00 | 0.02 | 1.03 | 0.36 |
| Bacteroidetes | Chryseobacterium | 224 | 68 | 5 | 8 | 0.83 | 0.25 | 0.02 | 0.03 |
| Bacteroidetes | Flavobacterium | 76 | 181 | 752 | 321 | 0.28 | 0.67 | 2.79 | 1.19 |
| Bacteroidetes | Lutibacter | 0 | 5 | 293 | 366 | 0.00 | 0.02 | 1.09 | 1.36 |
| Bacteroidetes | Sunxiuqinia | 2 | 3 | 274 | 28 | 0.01 | 0.01 | 1.01 | 0.10 |
| Bacteroidetes | unclassified-Bacteroidetes | 13 | 31 | 1129 | 509 | 0.05 | 0.11 | 4.18 | 1.89 |
| Cyanobacteria/Chloroplast | Bacillariophyta | 4 | 43 | 328 | 33 | 0.01 | 0.16 | 1.21 | 0.12 |
| Firmicutes | Bacillus | 465 | 52 | 73 | 79 | 1.72 | 0.19 | 0.27 | 0.29 |
| Firmicutes | Exiguobacterium | 2401 | 1387 | 8 | 4 | 8.89 | 5.14 | 0.03 | 0.01 |
| Firmicutes | Paenibacillus | 21 | 792 | 4 | 1 | 0.08 | 2.93 | 0.01 | 0.00 |
| Firmicutes | Trichococcus | 5 | 0 | 1 | 0 | 0.02 | 0.00 | 0.00 | 0.00 |
| Ignavibacteriae | Ignavibacterium | 2 | 1 | 261 | 302 | 0.01 | 0.00 | 0.97 | 1.12 |
| Planctomycetes | Planctomyces | 4 | 13 | 3 | 3 | 0.01 | 0.05 | 0.01 | 0.01 |
| Proteobacteria | Acidovorax | 81 | 255 | 9 | 30 | 0.30 | 0.94 | 0.03 | 0.11 |
| Proteobacteria | Aeromonas | 658 | 1952 | 55 | 21 | 2.44 | 7.23 | 0.20 | 0.08 |
| Proteobacteria | Arcobacter | 5 | 1 | 0 | 1 | 0.02 | 0.00 | 0.00 | 0.00 |
| Proteobacteria | Arenimonas | 24 | 56 | 1350 | 1353 | 0.09 | 0.21 | 5.00 | 5.01 |
| Proteobacteria | Azoarcus | 27 | 7 | 311 | 339 | 0.10 | 0.03 | 1.15 | 1.26 |
| Proteobacteria | Azospira | 0 | 6 | 42 | 44 | 0.00 | 0.02 | 0.16 | 0.16 |
| Proteobacteria | Azospirillum | 12 | 4 | 41 | 9 | 0.04 | 0.01 | 0.15 | 0.03 |
| Proteobacteria | Bradyrhizobium | 10 | 39 | 23 | 20 | 0.04 | 0.14 | 0.09 | 0.07 |
| Proteobacteria | Dechloromonas | 37 | 69 | 85 | 61 | 0.14 | 0.26 | 0.31 | 0.23 |
| Proteobacteria | Geobacter | 0 | 1 | 335 | 106 | 0.00 | 0.00 | 1.24 | 0.39 |
| Proteobacteria | Halomonas | 74 | 129 | 3911 | 5044 | 0.27 | 0.48 | 14.49 | 18.68 |
| Proteobacteria | Hydrogenophaga | 31 | 44 | 492 | 752 | 0.11 | 0.16 | 1.82 | 2.79 |
| Proteobacteria | Hyphomicrobium | 2 | 6 | 23 | 41 | 0.01 | 0.02 | 0.09 | 0.15 |
| Proteobacteria | Limnobacter | 1 | 2 | 44 | 174 | 0.00 | 0.01 | 0.16 | 0.64 |
| Proteobacteria | Mesorhizobium | 4 | 11 | 41 | 20 | 0.01 | 0.04 | 0.15 | 0.07 |
| Proteobacteria | Paraperlucidibaca | 0 | 1 | 252 | 19 | 0.00 | 0.00 | 0.93 | 0.07 |
| Proteobacteria | Perlucidibaca | 10 | 7 | 664 | 790 | 0.04 | 0.03 | 2.46 | 2.93 |
| Proteobacteria | Porphyrobacter | 59 | 92 | 207 | 80 | 0.22 | 0.34 | 0.77 | 0.30 |
| Proteobacteria | Pseudomonas | 20,068 | 17,756 | 1402 | 2603 | 74.33 | 65.76 | 5.19 | 9.64 |
| Proteobacteria | Pseudoxanthomonas | 1 | 2 | 6 | 6 | 0.00 | 0.01 | 0.02 | 0.02 |
| Proteobacteria | Rhizobium | 163 | 272 | 50 | 48 | 0.60 | 1.01 | 0.19 | 0.18 |
| Proteobacteria | Rhodobacter | 44 | 73 | 253 | 176 | 0.16 | 0.27 | 0.94 | 0.65 |
| Proteobacteria | Rhodoferax | 39 | 78 | 2088 | 863 | 0.14 | 0.29 | 7.73 | 3.20 |
| Proteobacteria | Sandaracinobacter | 0 | 0 | 84 | 307 | 0.00 | 0.00 | 0.31 | 1.14 |
| Proteobacteria | Shewanella | 0 | 17 | 5 | 1 | 0.00 | 0.06 | 0.02 | 0.00 |
| Proteobacteria | Simplicispira | 13 | 19 | 522 | 393 | 0.05 | 0.07 | 1.93 | 1.46 |
| Proteobacteria | Stenotrophomonas | 0 | 4 | 1 | 2 | 0.00 | 0.01 | 0.00 | 0.01 |
| Proteobacteria | Sulfuritalea | 1 | 5 | 207 | 644 | 0.00 | 0.02 | 0.77 | 2.39 |
| Proteobacteria | Thauera | 1 | 4 | 94 | 179 | 0.00 | 0.01 | 0.35 | 0.66 |
| Proteobacteria | Thiobacillus | 1 | 0 | 367 | 463 | 0.00 | 0.00 | 1.36 | 1.71 |
| Proteobacteria | unclassified-Rhodobacteraceae | 66 | 165 | 594 | 496 | 0.24 | 0.61 | 2.20 | 1.84 |
| Proteobacteria | unclassified-Sphingomonadales | 27 | 43 | 202 | 352 | 0.10 | 0.16 | 0.75 | 1.30 |
| Proteobacteria | unclassified-Xanthomonadaceae | 8 | 15 | 219 | 855 | 0.03 | 0.06 | 0.81 | 3.17 |
| Nitrospirae | Nitrospira | 0 | 1 | 3 | 2 | 0.0000 | 0.0037 | 0.0111 | 0.0074 |
| Proteobacteria | unclassified-Nitrosomonadaceae | 1 | 2 | 24 | 9 | 0.0037 | 0.0074 | 0.0889 | 0.0333 |
References
- Vymazal, J. Constructed wetlands for treatment of industrial wastewaters: A review. Ecol. Eng. 2014, 73, 724–751. [Google Scholar] [CrossRef]
- Zhang, D.Q.; Jinadasa, K.; Gersberg, R.M.; Liu, Y.; Ng, W.J.; Tan, S.K. Application of constructed wetlands for wastewater treatment in developing countries—A review of recent developments (2000–2013). J. Environ. Manag. 2014, 141, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Chen, H. Surface-Flow Constructed Treatment Wetlands for Pollutant Removal: Applications and Perspectives. Wetlands 2011, 31, 805–814. [Google Scholar] [CrossRef]
- Kadlec, R.H.; Wallace, S. Treatment Wetlands, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Faulwetter, J.L.; Gagnon, V.; Sundberg, C.; Chazarenc, F.; Burr, M.D.; Brisson, J.; Camper, A.K.; Stein, O.R. Microbial processes influencing performance of treatment wetlands: A review. Ecol. Eng. 2009, 35, 987–1004. [Google Scholar] [CrossRef]
- Saeed, T.; Sun, G. A review on nitrogen and organics removal mechanisms in subsurface flow constructed wetlands: Dependency on environmental parameters, operating conditions and supporting media. J. Environ. Manag. 2012, 112, 429–448. [Google Scholar] [CrossRef]
- Fu, G.; Huangshen, L.; Guo, Z.; Zhou, Q.; Wu, Z. Effect of plant-based carbon sources on denitrifying microorganisms in a vertical flow constructed wetland. Bioresour. Technol. 2017, 224, 214–221. [Google Scholar] [CrossRef]
- Truu, M.; Juhanson, J.; Truu, J. Microbial biomass, activity and community composition in constructed wetlands. Sci. Total Environ. 2009, 407, 3958–3971. [Google Scholar] [CrossRef]
- Vymazal, J. Constructed wetlands for wastewater treatment: Five decades of expericence. Environ. Sci. Technol. 2011, 45, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Meng, P.; Pei, H.; Hu, W.; Shao, Y.; Li, Z. How to increase microbial degradation in constructed wetlands: Influencing factors and improvement measures. Bioresour. Technol. 2014, 157, 316–326. [Google Scholar] [CrossRef]
- Vymazal, J.; Kröpfelová, L. Removal of organics in constructed wetlands with horizontal sub-surface flow: A review of the field experience. Sci. Total Environ. 2009, 407, 3911–3922. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, D.; Horton, D.; Griffin, P.; Vanrolleghem, P.; De Pauw, N. Impact of operational maintenance on the asset life of storm reed beds. Water Sci. Technol. 2005, 51, 243–250. [Google Scholar] [CrossRef]
- Faulwetter, J.L.; Burr, M.D.; Parker, A.E.; Stein, O.R.; Camper, A.K. Influence of season and plant species on the abundance and diversity of sulfate reducing bacteria and ammonia oxidizing bacteria in constructed wetland microcosms. Microb. Ecol. 2013, 65, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Shelef, O.; Gross, A.; Rachmilevitch, S. Role of Plants in a Constructed Wetland: Current and New Perspectives. Water 2013, 5, 405–419. [Google Scholar] [CrossRef]
- Ahn, C.; Gillevet, P.; Sikaroodi, M. Molecular characterization of microbial communities in treatment microcosm wetlands as influenced by macrophytes and phosphorus loading. Ecol. Indic. 2007, 7, 852–863. [Google Scholar] [CrossRef]
- Gorra, R.; Coci, M.; Ambrosoli, R.; Laanbroek, H. Effects of substratum on the diversity and stability of ammonia-oxidizing communities in a constructed wetland used for wastewater treatment. J. Appl. Microbiol. 2007, 103, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- DeJournett, T.D.; Arnold, W.A.; LaPara, T.M. The characterization and quantification of methanotrophic bacterial populations in constructed wetland sediments using PCR targeting 16S rRNA gene fragments. Appl. Soil Ecol. 2007, 35, 648–659. [Google Scholar] [CrossRef]
- Baptista, J.D.C.; Davenport, R.J.; Donnelly, T.; Curtis, T.P. The microbial diversity of laboratory-scale wetlands appears to be randomly assembled. Water Res. 2008, 42, 3182–3190. [Google Scholar] [CrossRef] [PubMed]
- Brisson, J.; Chazarenc, F. Maximizing pollutant removal in constructed wetlands: Should we pay more attention to macrophyte species selection? Sci. Total Environ. 2009, 407, 3923–3930. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, P.; Anderson, J.C.; Carlson, J.C.; Low, J.E.; Challis, J.K.; Beattie, S.A.; Bartel, C.N.; Elliott, A.D.; Montero, O.F.; Lokesh, S.; et al. Macrophytes may not contribute significantly to removal of nutrients, pharmaceuticals, and antibiotic resistance in model surface constructed wetlands. Sci. Total Environ. 2014, 482, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, J. Improving Winter Performance of Constructed Wetlands for Wastewater Treatment in Northern China: A Review. Wetlands 2013, 34, 243–253. [Google Scholar] [CrossRef]
- Gao, J.; Wang, W.; Guo, X.; Zhu, S.; Chen, S.; Zhang, R. Nutrient removal capability and growth characteristics of Iris sibirica in subsurface vertical flow constructed wetlands in winter. Ecol. Eng. 2014, 70, 351–361. [Google Scholar] [CrossRef]
- Zou, X.; Zhang, H.; Zuo, J.; Wang, P.; Zhao, D.; An, S. Decreasing but still significant facilitation effect of cold-season macrophytes on wetlands purification function during cold winter. Sci. Rep. 2016, 6, 27011. [Google Scholar] [CrossRef] [PubMed]
- Vymazal, J. Emergent plants used in free water surface constructed wetlands: A review. Ecol. Eng. 2013, 61, 582–592. [Google Scholar] [CrossRef]
- Hang, Q.; Wang, H.; Chu, Z.; Ye, B.; Li, C.; Hou, Z. Application of plant carbon source for denitrification by constructed wetland and bioreactor: Review of recent development. Environ. Sci. Pollut. Res. 2016, 23, 8260–8274. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, G. Nutrient concentration variations during Oenanthe javanica growth and decay in the ecological floating bed system. J. Environ. Sci. 2010, 22, 1710–1717. [Google Scholar] [CrossRef]
- Chen, Z.; Cuervo, D.P.; Müller, J.A.; Wiessner, A.; Köser, H.; Vymazal, J.; Kästner, M.; Kuschk, P. Hydroponic root mats for wastewater treatment—A review. Environ. Sci. Pollut. Res. 2016, 23, 15911–15928. [Google Scholar] [CrossRef]
- Rice, E.; Baird, R.; Eaton, A.; Clesceri, L. Standard Methods for the Examination of Water and Waste Water; American Public Health Association: Washington, DC, USA, 2012. [Google Scholar]
- Li, H.S.; Sun, Q.; Zhao, S.J.; Zhang, W.H. Principles and Techniques of Pant Physiological Biochemical Experiment; Higher Education Press: Beiijng, China, 2000. [Google Scholar]
- Toet, S.; Bouwman, M.; Cevaal, A.; Verhoeven, J.T.A. Nutrient removal through autumn harvest ofphragmites australisandthypha latifoliashoots in relation to nutrient loading in a wetland system used for polishing sewage treatment plant effluent. Environ. Lett. 2005, 40, 1133–1156. [Google Scholar] [CrossRef]
- Kludze, H.K.; Delaune, R.D.; Patrick, W.H. A Colorimetric Method for Assaying Dissolved Oxygen Loss from Container-Grown Rice Roots. Agron. J. 1994, 86, 483–487. [Google Scholar] [CrossRef]
- Wu, C.; Shu, W.; Zhu, Y.-G.; Ye, Z.; Wong, M. Arsenic accumulation and speciation in rice are affected by root aeration and variation of genotypes. J. Exp. Bot. 2011, 62, 2889–2898. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wen, Y.; Zhou, Q.; Vymazal, J. Effects of plant biomass on denitrifying genes in subsurface-flow constructed wetlands. Bioresour. Technol. 2014, 157, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef]
- Peet, R.K. The Measurement of Species Diversity. Annu. Rev. Ecol. Syst. 1974, 5, 285–307. [Google Scholar] [CrossRef]
- Heylen, K.; Vanparys, B.; Wittebolle, L.; Verstraete, W.; Boon, N.; De Vos, P. Cultivation of Denitrifying Bacteria: Optimization of Isolation Conditions and Diversity Study. Appl. Environ. Microbiol. 2006, 72, 2637–2643. [Google Scholar] [CrossRef]
- Philippot, L.; Hallin, S.; Schloter, M. Ecology of Denitrifying Prokaryotes in Agricultural Soil. Adv. Agron. 2007, 96, 249–305. [Google Scholar]
- Wu, S.; Kuschk, P.; Brix, H.; Vymazal, J.; Dong, R. Development of constructed wetlands in performance intensifications for wastewater treatment: A nitrogen and organic matter targeted review. Water Res. 2014, 57, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wallace, S.; Brix, H.; Kuschk, P.; Kirui, W.K.; Masi, F.; Dong, R. Treatment of industrial effluents in constructed wetlands: Challenges, operational strategies and overall performance. Environ. Pollut. 2015, 201, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, C.; Ji, G.; Zhi, W.; Sheng, L. Nitrogen removal pathways in a tidal flow constructed wetland under flooded time constraints. Ecol. Eng. 2015, 81, 266–271. [Google Scholar] [CrossRef]
- Cui, L.; Ouyang, Y.; Gu, W.; Yang, W.; Xu, Q. Evaluation of nutrient removal efficiency and microbial enzyme activity in a baffled subsurface-flow constructed wetland system. Bioresour. Technol. 2013, 146, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Wu, J.; Dai, Y.; Yang, L.; Zhang, Z.; Cheng, S.; Zhang, Q. Bacterial community analysis by pcr-dgge and 454-pyrosequencing of horizontal subsurface flow constructed wetlands with front aeration. Appl. Microbiol. Biotechnol. 2015, 99, 1499–1512. [Google Scholar] [CrossRef]
- Saunders, A.M.; Larsen, P.; Nielsen, P.H. Comparison of nutrient-removing microbial communities in activated sludge from full-scale MBRs and conventional plants. Water Sci. Technol. 2013, 68, 366–371. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, Y.; Tang, Z.; Huang, J.; Zhou, Q.; Vymazal, J. Effects of plant biomass on bacterial community structure in constructed wetlands used for tertiary wastewater treatment. Ecol. Eng. 2015, 84, 38–45. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, H.; Ngo, H.H.; Guo, W.; Zhang, J.; Liu, C.; Liang, S.; Hu, Z.; Yang, Z.; Zhao, C. Microbial abundance and community in subsurface flow constructed wetland microcosms: Role of plant presence. Environ. Sci. Pollut. Res. Int. 2016, 23, 4036–4045. [Google Scholar] [CrossRef] [PubMed]
- Salvato, M.; Borin, M.; Doni, S.; Macci, C.; Ceccanti, B.; Marinari, S.; Masciandaro, G. Wetland plants, micro-organisms and enzymatic activities interrelations in treating N polluted water. Ecol. Eng. 2012, 47, 36–43. [Google Scholar] [CrossRef]
- Menon, R.; Jackson, C.R.; Holland, M.M. The Influence of Vegetation on Microbial Enzyme Activity and Bacterial Community Structure in Freshwater Constructed Wetland Sediments. Wetlands 2013, 33, 365–378. [Google Scholar] [CrossRef]
- Sollai, M.; Hopmans, E.C.; Schouten, S.; Keil, R.G.; Damste, J.S.S. Intact polar lipids of Thaumarchaeota and anammox bacteria as indicators of N-cycling in the Eastern Tropical North Pacific oxygen deficient zone. Biogeosci. Discuss. 2015, 12, 4833–4864. [Google Scholar] [CrossRef]
- Paranychianakis, N.V.; Tsiknia, M.; Kalogerakis, N. Pathways regulating the removal of nitrogen in planted and unplanted subsurface flow constructed wetlands. Water Res. 2016, 102, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Berg, C.; Vandieken, V.; Thamdrup, B.; Jurgens, K. Significance of archaeal nitrification in hypoxic waters of the baltic sea. ISME J. 2015, 9, 1319–1332. [Google Scholar] [CrossRef]
- Zhi, W.; Ji, G. Quantitative response relationships between nitrogen transformation rates and nitrogen functional genes in a tidal flow constructed wetland under C/N ratio constraints. Water Res. 2014, 64, 32–41. [Google Scholar] [CrossRef]
- Ye, F.; Li, Y. Enhancement of nitrogen removal in towery hybrid constructed wetland to treat domestic wastewater for small rural communities. Ecol. Eng. 2009, 35, 1043–1050. [Google Scholar] [CrossRef]
- Wang, W.; Ding, Y.; Ullman, J.L.; Ambrose, R.F.; Song, X.; Zhao, Z. Nitrogen removal performance in planted and unplanted horizontal subsurface flow constructed wetlands treating different influent COD/N ratios. Environ. Sci. Pollut. Res. 2016, 23, 9012–9018. [Google Scholar] [CrossRef] [PubMed]
- Vymazal, J. Removal of nutrients in various types of constructed wetlands. Sci. Total Environ. 2007, 380, 48–65. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, J.; Jia, W.; Xie, H.; Gu, R.R.; Li, C.; Gao, B. Impact of COD/N ratio on nitrous oxide emission from microcosm wetlands and their performance in removing nitrogen from wastewater. Bioresour. Technol. 2009, 100, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Jetten, M.S.M.; Kuschk, P.; Ettwig, K.F.; Yin, C. Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl. Microbiol. Biotechnol. 2010, 86, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.S.; Kuenen, G.; Wagner, M.; Fuerst, J.; Van Loosdrecht, M.; Strous, M. Microbiology and application of the anaerobic ammonium oxidation (‘anammox’) process. Curr. Opin. Biotechnol. 2001, 12, 283–288. [Google Scholar] [CrossRef]
- Ansola, G.; Arroyo, P.; De Miera, L.E.S. Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Sci. Total Environ. 2014, 473, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Q.; Fan, J.; Xie, H.; Liu, C.; Liang, S.; Hu, Z.; Yang, Z.; Zhao, C. Comparisons of microbial abundance and community among different plant species in constructed wetlands in summer. Ecol. Eng. 2015, 82, 376–380. [Google Scholar] [CrossRef]
- Lv, X.; Yu, J.; Fu, Y.; Ma, B.; Qu, F.; Ning, K.; Wu, H. A Meta-Analysis of the Bacterial and Archaeal Diversity Observed in Wetland Soils. Sci. J. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Liu, S.; Ying, G.-G.; Liu, Y.-S.; Peng, F.-Q.; He, L.-Y. Degradation of Norgestrel by Bacteria from Activated Sludge: Comparison to Progesterone. Environ. Sci. Technol. 2013, 47, 10266–10276. [Google Scholar] [CrossRef]
- Nakaya, A.; Onodera, Y.; Nakagawa, T.; Satoh, K.; Takahashi, R.; Sasaki, S.; Tokuyama, T. Analysis of Ammonia Monooxygenase and Archaeal 16S rRNA Gene Fragments in Nitrifying Acid-Sulfate Soil Microcosms. Microbes Environ. 2009, 24, 168–174. [Google Scholar] [CrossRef]
- Tsushima, I.; Kindaichi, T.; Okabe, S. Quantification of anaerobic ammonium-oxidizing bacteria in enrichment cultures by real-time PCR. Water Res. 2007, 41, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Okano, Y.; Hristova, K.R.; Leutenegger, C.M.; Jackson, L.E.; Denison, R.F.; Gebreyesus, B.; Lebauer, D.; Scow, K.M. Application of Real-Time PCR to Study Effects of Ammonium on Population Size of Ammonia-Oxidizing Bacteria in Soil. Appl. Environ. Microbiol. 2004, 70, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Scala, D.J.; Kerkhof, L.J. Nitrous oxide reductase (nosz) gene-specific pcr primers for detection of denitrifiers and three nosz genes from marine sediments. FEMS Microbiol. Lett. 1998, 162, 61–68. [Google Scholar] [CrossRef]
- Throback, I.N.; Enwall, K.; Jarvis, A.; Hallin, S. Reassessing pcr primers targeting nirs, nirk and nosz genes for community surveys of denitrifying bacteria with dgge. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef]
- Yan, T.; Zu, Y.; Fields, M.W.; Tiedje, J.M.; Wu, L.; Zhou, J. Molecular diversity and characterization of nitrite reductase gene fragments (nirk and nirs) from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 2003, 5, 13–24. [Google Scholar] [CrossRef]


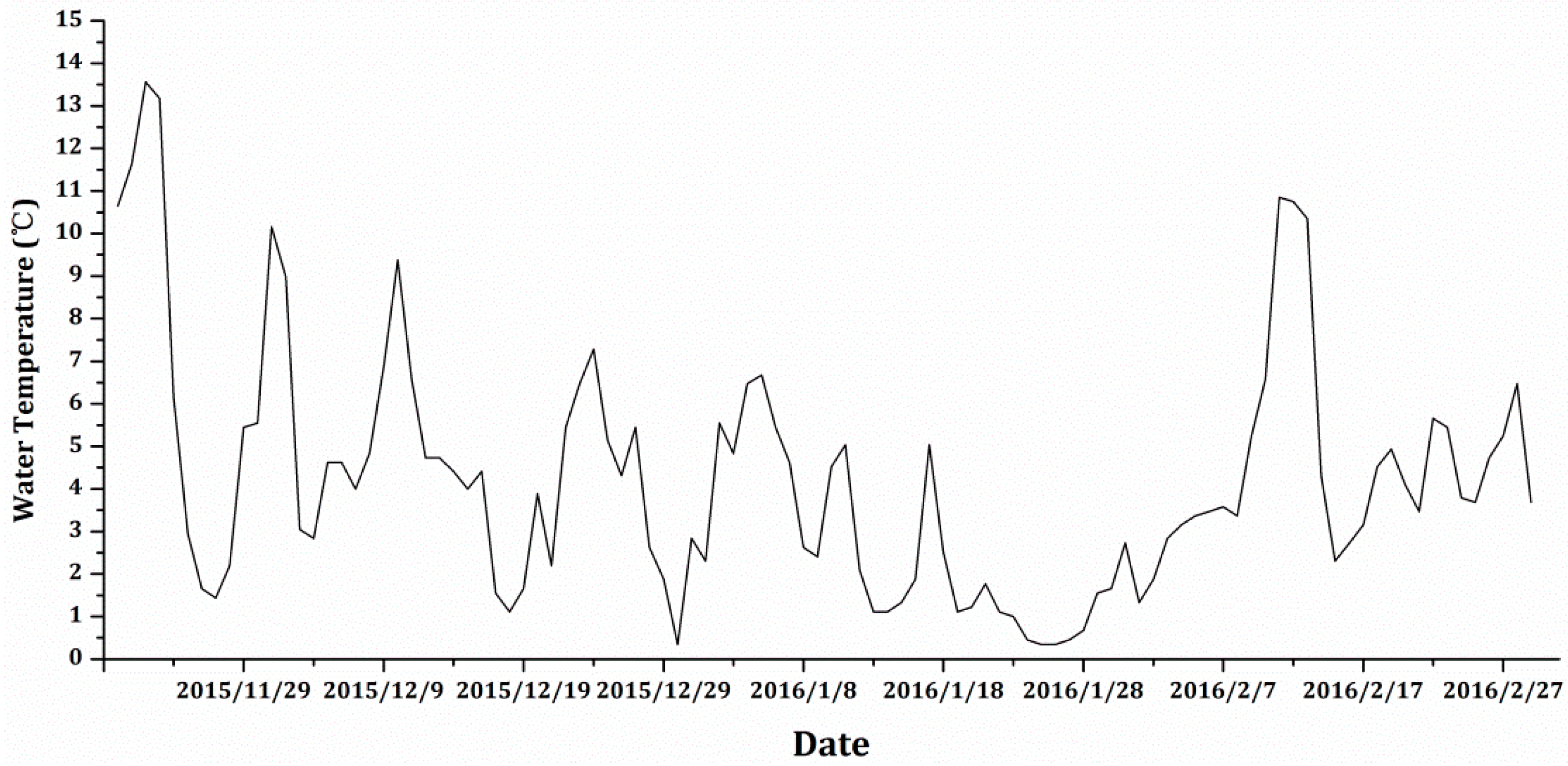
| Parameter | 20 December 2015 | 19 January 2016 | 29 January 2016 | 18 February 2016 |
|---|---|---|---|---|
| NH4+–N (mg·L−1) | 8.9 ± 0.9 | 10.8 ± 0.4 | 10.8 ± 0.3 | 11.6 ± 0.5 |
| NO3−–N (mg·L−1) | 10.5 ± 0.8 | 10.8 ± 0.7 | 10..8 ± 0.9 | 10.4 ± 0.4 |
| NO2−–N (mg·L−1) | 0.650 ± 0.025 | 0.560 ± 0.025 | 0.380 ± 0.005 | 0.335 ± 0.005 |
| TN (mg·L−1) | 24.8 ± 1.5 | 25.7 ± 3.0 | 25.3 ± 0.9 | 26.6 ± 2.0 |
| COD (mg·L−1) | 14.4 ± 2.2 | 15.4 ± 2.5 | 14.6 ± 2.8 | 15.8 ± 1.8 |
| DO (mg·L−1) | 9.7 ± 0.8 | 8.9 ± 0.5 | 9.3 ± 0.9 | 8.3 ± 0.6 |
| pH | 7.88 ± 0.65 | 7.05 ± 0.45 | 6.45 ± 0.98 | 8.45 ± 1.24 |
| Phase | System | Shoot Length (cm) | Root Length (cm) | Shoot Biomass (g·m−2) | Root Biomass (g·m−2) | N in Shoot (g·m−2) | N in Root (g·m−2) | Root Activity (μg TTC·g−1 Root·h−1) | ROL Rate (μmol O2·g−1 Root·h−1) |
|---|---|---|---|---|---|---|---|---|---|
| Initial | Tcw | 50.0 ± 2.5 | 15.0 ± 1.0 | 95.0 ± 6.3 | 35.0 ± 3.6 | 1.425 | 0.315 | 48.5 ± 6.4 | 0.96 ± 0.045 |
| Tcp | 50.0 ± 2.5 | 15.0 ± 1.0 | 95.0 ± 6.3 | 35.0 ± 3.6 | 1.425 | 0.315 | 48.5 ± 6.4 | 0.96 ± 0.045 | |
| 2nd Batch | Tcw | 52.0 ± 2.7 | 16.0 ± 1.4 | 86.0 ± 5.3 | 35.8 ± 5.5 | 1.290 | 0.322 | 42.6 ± 3.7 | 0.68 ± 0.097 |
| Tcp | 52.0 ± 2.8 | 16.0 ± 1.3 | 84.0 ± 6.5 | 36.4 ± 3.7 | 1.260 | 0.328 | 46.7 ± 5.2 | 0.88 ± 0.098 | |
| 4th Batch | Tcw | 55.0 ± 3.5 | 17.0 ± 1.5 | 87.0 ± 5.8 | 37.0 ± 4.1 | 1.305 | 0.333 | 43.2 ± 5.3 | 0.79 ± 0.054 |
| Tcp | 55.0 ± 3.4 | 17.7 ± 1.2 | 87.2 ± 7.6 | 38.3 ± 3.3 | 1.305 | 0.345 | 46.4 ± 5.5 | 0.87 ± 0.093 | |
| 6th Batch | Tcw | 54.0 ± 2.6 | 20.5 ± 2.0 | 92.3 ± 5.7 | 40.5 ± 5.2 | 1.385 | 0.365 | 42.8 ± 4.5 | 1.02 ± 0.065 |
| Tcp | 53.0 ± 1.7 | 20.0 ± 2.4 | 91.6 ± 6.5 | 40.6 ± 5.1 | 1.374 | 0.365 | 47.5 ± 6.2 | 1.03 ± 0.061 | |
| 8th Batch | Tcw | 56.0 ± 2.5 | 22.6 ± 2.2 | 102.3 ± 13.5 | 46.8 ± 4.3 | 1.535 | 0.421 | 58.7 ± 6.5 | 1.28 ± 0.078 |
| Tcp | 55.0 ± 2.5 | 23.0 ± 2.7 | 104.5 ± 14.4 | 48.0 ± 4.8 | 1.568 | 0.432 | 63.6 ± 8.9 | 1.48 ± 0.085 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Wang, P.; Liu, Y.; Zhao, D.; Leng, X.; An, S. Effects of Oenanthe javanica on Nitrogen Removal in Free-Water Surface Constructed Wetlands under Low-Temperature Conditions. Int. J. Environ. Res. Public Health 2019, 16, 1420. https://doi.org/10.3390/ijerph16081420
Song S, Wang P, Liu Y, Zhao D, Leng X, An S. Effects of Oenanthe javanica on Nitrogen Removal in Free-Water Surface Constructed Wetlands under Low-Temperature Conditions. International Journal of Environmental Research and Public Health. 2019; 16(8):1420. https://doi.org/10.3390/ijerph16081420
Chicago/Turabian StyleSong, Siyuan, Penghe Wang, Yongxia Liu, Dehua Zhao, Xin Leng, and Shuqing An. 2019. "Effects of Oenanthe javanica on Nitrogen Removal in Free-Water Surface Constructed Wetlands under Low-Temperature Conditions" International Journal of Environmental Research and Public Health 16, no. 8: 1420. https://doi.org/10.3390/ijerph16081420





