Converging Human and Malaria Vector Diagnostics with Data Management towards an Integrated Holistic One Health Approach
Abstract
:1. Introduction
2. Diagnostics
2.1. State-of-the-Art and Challenges in Human Diagnostics
2.2. State-of-the-Art and Challenges in Vector Diagnostics
2.3. Progress beyond the State-of-the-Art
- They offer the capability of multiplexing, thus increasing throughput without the need of costly and complex analysis methodologies, such as microarrays.
- They are diagnostically robust, they offer high sensitivity and specificity, they have been introduced in routine laboratory use for years, and are applicable to most laboratory settings.
- The exact same assays that are used in human diagnostics for pathogen detection can also be used in vector diagnostics (exact same species/serotypes/lineages of pathogens can be detected), thus making epidemiological information more universal.
- Both DNA and RNA determinations can be performed with exactly the same methodology from sample preparation to measurement.
- Especially TaqMan PCR allows (semi)quantitative information about the degree of infection in humans, enabling a correlation between the Ct value and the pathogen concentration (parasite/bacterium/virus).
- They can be integrated in an automated platform.
3. Packaging into Portable and Automated Platforms
3.1. Automation and Integration
- Provide the suitable interface for safe and contamination-free sample inlet. The sample inlet should be of universal nature in order to facilitate analysis from different matrices, such as whole blood, serum, sputum (in case of tuberculosis), or swab transport medium (in case of respiratory infections), but also mosquito homogenate samples.
- Automate sample preparation and interface it on the same cartridge with the NAAT mentioned in Section 2.
- Integrate and in situ store (bio)chemical reagents that are required for sample-to-answer analysis.
- Use a processing instrument, which incorporates the detection technology and reporting interface.
- Be adaptable in exchanging the desired targets according to endemic and epidemic needs, and geographical requirements.
3.2. State-of-the-Art in Automated Detection Technologies
3.2.1. VerePLEXTM Biosystem
3.2.2. Q-POCTM
3.2.3. LabDisk
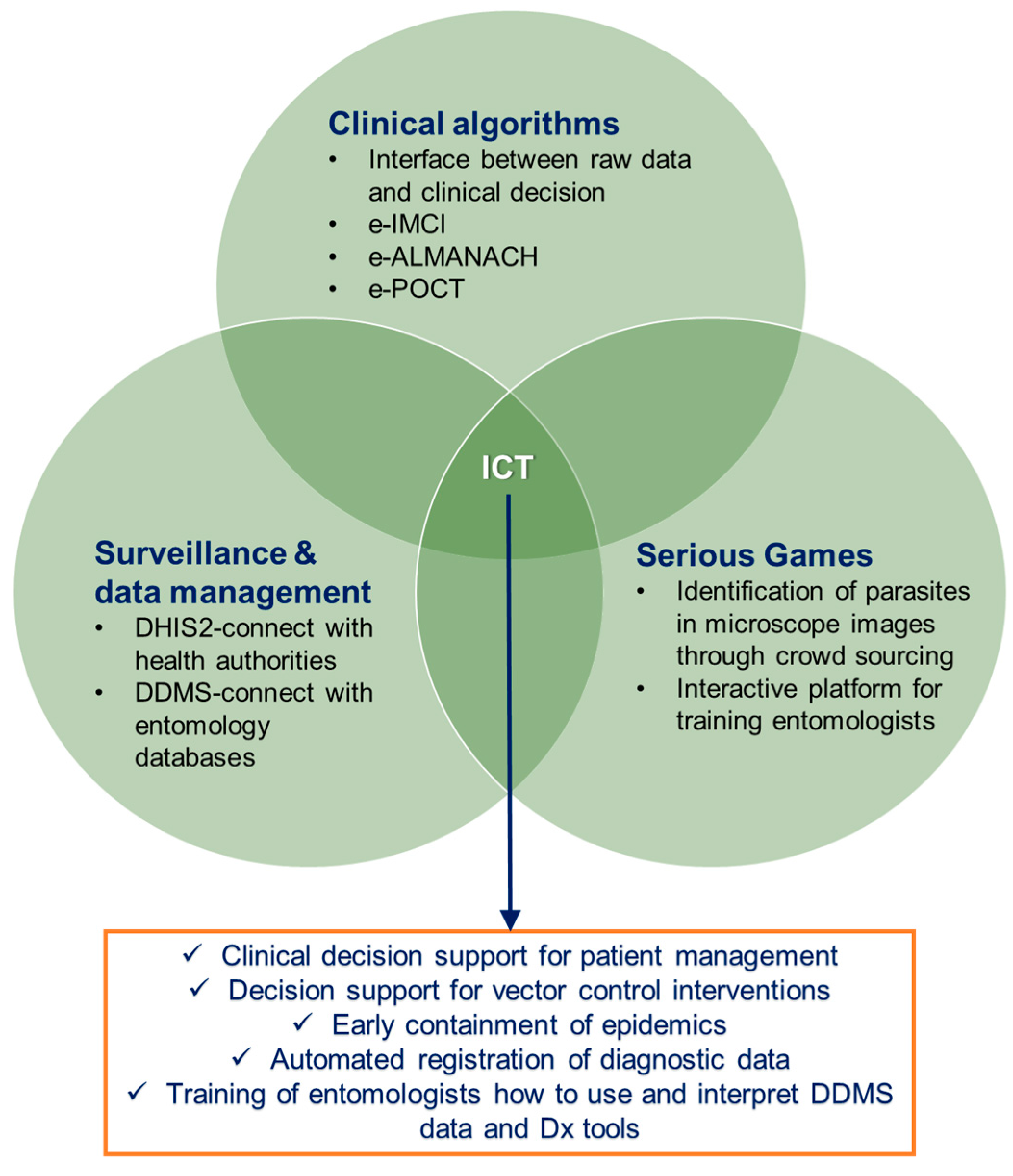
4. Inclusion of Information and Communication Technologies
4.1. Clinical Algorithms for Patient Management
4.2. Surveillance and Data Management
4.2.1. Surveillance in Human Diagnostics
4.2.2. Data Management Systems in Entomology and Modern Communication Tools
4.2.3. An Integrated Data Management and Decision-Support System
4.3. Serious Games
5. Behavioral Change and Impact of a Holistic Approach
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO. World Malaria Report; World Health Organization: Geneva, Switzerland, 2017; ISBN 9789241565523. [Google Scholar]
- Kaga, T.; Ohta, S. Ecophysiological and Climatological Effects on Distribution of Vector Species and Malaria Incidence in India. Int. J. Environ. Res. Public Health 2012, 9, 4704–4714. [Google Scholar] [CrossRef] [PubMed]
- Piperaki, E.T.; Daikos, G.L. Malaria in Europe: Emerging threat or minor nuisance? Clin. Microbiol. Infect. 2016, 22, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Chang, H.L.; Chen, K.T. The Epidemiology of Imported Malaria in Taiwan between 2002-2013: The Importance of Sensitive Surveillance and Implications for Pre-Travel Medical Advice. Int. J. Environ. Res. Public Health 2014, 11, 5651–5664. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.F.; Delage, G.; Seed, C.R.; Pillonel, J.; Fabra, C.C.; Davison, K.; Kitchen, A.; Steele, W.R.; Leiby, D.A. The Epidemiology of Imported Malaria and Transfusion Policy in 5 Nonendemic Countries. Transfus. Med. Rev. 2015, 29, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Lo Re, V.; Gluckman, S.J. Fever in the Returned Traveler. Am. Fam. Physician 2003, 68, 1343–1350. [Google Scholar] [PubMed]
- Tatem, A.J.; Jia, P.; Ordanovich, D.; Falkner, M.; Huang, Z.; Howes, R.; Hay, S.I.; Gething, P.W.; Smith, D.L. The geography of imported malaria to non-endemic countries: A meta-analysis of nationally reported statistics. Lancet Infect. Dis. 2017, 17, 98–107. [Google Scholar] [CrossRef]
- WHO. Global Technical Strategy for Malaria 2016–2030; World Health Organization: Geneva, Switzerland, 2015; ISBN 9789241564991. Available online: http://www.who.int/malaria/publications/atoz/9789241564991/en/ (accessed on 21 January 2018).
- Molineaux, L.; Gramiccia, G. The Garki Project: Research on the Epidemiology and Control of Malaria in the Sudan Savanna of West Africa; World Health Organization: Geneva, Switzerland, 1980. [Google Scholar]
- Nájera, J.A.; González-Silva, M.; Alonso, P.L. Some lessons for the future from the global malaria eradication programme (1955–1969). PLoS Med. 2011, 8, e1000412. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.A.; Mboera, L.E.G.; Senkoro, K.; Lesser, A.; Shayo, E.H.; Paul, C.J.; Miranda, M.L. A Randomized Longitudinal Factorial Design to Assess Malaria Vector Control and Disease Management Interventions in Rural Tanzania. Int. J. Environ. Res. Public Health 2014, 11, 5317–5332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, B.; Kahn, L.H.; Monath, T.P.; Woodall, J. ‘ONE HEALTH’ and parasitology. Parasite Vector 2009, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- One Health Initiative Will Unite Human and Veterinary Medicine. Available online: http://www.onehealthinitiative.com/ (accessed on 11 November 2017).
- WHO. Guidelines for the Treatment of Malaria, 3rd ed.; World Health Organization: Geneva, Switzerland, 2015; ISBN 9789241549127. [Google Scholar]
- WHO. Emergency Response to Artemisinin Resistance in the Greater Mekong Subregion: Regional Framework for Action 2013–2015; World Health Organization: Geneva, Switzerland, 2013; ISBN 9789241505321. Available online: http://www.who.int/malaria/publications/atoz/9789241505321/en/ (accessed on 8 November 2017).
- D’Acremont, V.; Kilowoko, M.; Kyungu, E.; Philipina, S.; Sangu, W.; Kahama-Maro, J.; Lengeler, C.; Cherpillod, P.; Kaiser, L.; Genton, B. Beyond Malaria-Causes of Fever in Outpatient Tanzanian Children. N. Engl. J. Med. 2014, 370, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Morrissey, A.B.; Nicholson, W.L.; Massung, R.F.; Stoddard, R.A.; Galloway, R.L.; Ooi, E.E.; Maro, V.P.; Saganda, W.; Kinabo, G.D.; et al. Etiology of Severe Non-malaria Febrile Illness in Northern Tanzania: A Prospective Cohort Study. PLoS Neglect. Trop. Dis. 2013, 7, e2324. [Google Scholar] [CrossRef] [PubMed]
- Makani, J.; Matuja, W.; Liyombo, E.; Snow, R.W.; Marsh, K.; Warrell, D.A. Admission diagnosis of cerebral malaria in adults in an endemic area of Tanzania: Implications and clinical description. QJM-Int. J. Med. 2003, 96, 355–362. [Google Scholar] [CrossRef]
- Reyburn, H.; Mbatia, R.; Drakeley, C.; Carneiro, I.; Mwakasungula, E.; Mwerinde, O.; Saganda, K.; Shao, J.; Kitua, A.; Olomi, R.; et al. Overdiagnosis of malaria in patients with severe febrile illness in Tanzania: A prospective study. Br. Med. J. 2004, 329, 1212–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biggs, H.M.; Lester, R.; Nadjm, B.; Mtove, G.; Todd, J.E.; Kinabo, G.D.; Philemon, R.; Amos, B.; Morrissey, A.B.; Reyburn, H.; et al. Invasive Salmonella Infections in Areas of High and Low Malaria Transmission Intensity in Tanzania. Clin. Infect. Dis. 2014, 58, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Boillat, N.; Josephine, S.; Mbarack, Z.; Mlaganile, T.; Kazimoto, T.; Genton, B.; D’Acremont, V. Etiologies of acute febrile illness among adults attending outpatient clinics in Dar es Salaam, Tanzania. In Proceedings of the American Society of Tropical Medicine and Hygiene (ASTMH), New Orleans, LA, USA, 2–6 November 2014; Late breakers Abstract book, LB-3155. pp. 322–323. [Google Scholar]
- Kelland, K. Global Health Experts Accuse WHO of “Egregious Failure” on Ebola. Clin. Infect. Dis. 2014, 59, i–ii. [Google Scholar]
- D’Acremont, V.; Kahama-Maro, J.; Swai, N.; Mtasiwa, D.; Genton, B.; Lengeler, C. Reduction of anti-malarial consumption after rapid diagnostic tests implementation in Dar es Salaam: A before-after and cluster randomized controlled study. Malar. J. 2011, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Lubell, Y.; Staedke, S.G.; Greenwood, B.M.; Kamya, M.R.; Molyneux, M.; Newton, P.N.; Reyburn, H.; Snow, R.W.; D’Alessandro, U.; English, M.; et al. Likely Health Outcomes for Untreated Acute Febrile Illness in the Tropics in Decision and Economic Models; A Delphi Survey. PLoS ONE 2011, 6, e17439. [Google Scholar] [CrossRef] [PubMed]
- Tangpukdee, N.; Duangdee, C.; Wilairatana, P.; Krudsood, S. Malaria Diagnosis: A Brief Review. Korean J. Parasitol. 2009, 47, 93–102. [Google Scholar] [CrossRef] [PubMed]
- WHO. Microscopy; World Health Organization: Geneva, Switzerland, 2017; Available online: www.who.int/malaria/areas/diagnosis/microscopy/en (accessed on 9 November 2017).
- Cheng, Q.; Cunningham, J.; Gatton, M.L. Systematic Review of Sub-microscopic P. vivax Infections: Prevalence and Determining Factors. PLoS Neglect. Trop. Dis. 2015, 9, e3413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okell, L.C.; Ghani, A.C.; Lyons, E.; Drakeley, C.J. Submicroscopic infection in Plasmodium falciparum-endemic populations: A systematic review and meta-analysis. J. Infect. Dis. 2009, 200, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- WHO/FIND/CDC. Malaria Rapid Diagnostic Test Performance: Summary Results of WHO Product Testing of Malaria RDTs: Rounds 1-6 (2008–2015); World Health Organization: Geneva, Switzerland, 2015; ISBN 9789241510042. Available online: http://apps.who.int/iris/bitstream/10665/204119/1/9789241510042_eng.pdf (accessed on 3 November 2017).
- Incardona, S.; Serra-Casas, E.; Champouillon, N.; Nsanzabana, C.; Cunningham, J.; González, I.J. Global survey of malaria rapid diagnostic test (RDT) sales, procurement and lot verification practices: Assessing the use of the WHO–FIND Malaria RDT Evaluation Programme (2011–2014). Malar. J. 2017, 16, 196. [Google Scholar] [CrossRef] [PubMed]
- Kosasih, H.; Widjaja, S.; Surya, E.; Hadiwijaya, S.H.; Butarbutar, D.P.; Jaya, U.A.; Nurhayati, A.B.; Williams, M. Evaluation of two IgM rapid immunochromatographic tests during circulation of Asian lineage Chikungunya virus. Southeast Asian J. Trop. Med. Public Health 2012, 43, 55–61. [Google Scholar] [PubMed]
- Cook, J.; Xu, W.; Msellem, M.; Vonk, M.; Bergstrom, B.; Gosling, R.; Al-Mafazy, A.W.; McElroy, P.; Molteni, F.; Abass, A.K.; et al. Mass screening and treatment on the basis of results of a Plasmodium falciparum-specific rapid diagnostic test did not reduce malaria incidence in Zanzibar. J. Infect. Dis. 2015, 211, 1476–1483. [Google Scholar] [CrossRef] [PubMed]
- UNITAID. Malaria Diagnostics Technology and Market Landscape Report, 3rd ed.; UNITAID: Geneva, Switzerland, 2016; Available online: https://www.ghdonline.org/uploads/Unitaid-Malaria-Dx-Tech-Mkt-Landscape-3rd-Ed-April-2016.pdf (accessed on 9 November 2017).
- Britton, S.; Cheng, Q.; McCarthy, J.S. Novel molecular diagnostic tools for malaria elimination: A review of options from the point of view of high-throughput and applicability in resource limited settings. Malar. J. 2016, 15, 88. [Google Scholar] [CrossRef] [PubMed]
- Cordray, M.S.; Richards-Kortum, R.R. Review: Emerging Nucleic Acid Based Tests for Point-of-Care Detection of Malaria. Am. J. Trop. Med. Hyg. 2012, 7, 223–230. [Google Scholar] [CrossRef] [PubMed]
- WHO. Policy Brief on Malaria Diagnostic in Low Transmission Settings; World Health Organization: Geneva, Switzerland, 2014; Available online: http://www.who.int/malaria/publications/atoz/policy-brief-diagnosis-low-transmission-settings/en/ (accessed on 9 December 2017).
- Vo, T.K.; Bigot, P.; Gazin, P.; Sinou, V.; De Pina, J.J.; Huynh, D.C.; Fumoux, F.; Parzy, D. Evaluation of a real-time PCR assay for malaria diagnosis in patients from Vietnam and in returned travelers. Trans. R. Soc. Trop. Med. 2007, 101, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Mens, P.F.; Schoone, G.J.; Kager, P.A.; Schallig, H.D. Detection and identification of human Plasmodium species with real time quantitative nucleic acid sequence based amplification. Malar. J. 2006, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Safeukui, I.; Millet, P.; Boucher, S.; Melinard, L.; Fregeville, F.; Receveur, M.C.; Pistone, T.; Fialon, P.; Vincendeau, P.; Fleury, H.; et al. Evaluation of FRET real-time PCR assay for rapid detection and differentiation of Plasmodium species in returning travellers and migrants. Malar. J. 2008, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Perandin, F.; Manca, N.; Calderaro, A.; Piccolo, G.; Galati, L.; Ricci, L.; Medici, M.C.; Arcangeletti, M.C.; Snounou, G.; Dettori, G.; et al. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J. Clin. Microbiol. 2004, 42, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Mangold, K.A.; Manson, R.U.; Koay, E.S.C.; Stephens, L.; Regner, M.; Thomson, R.B.; Peterson, L.R.; Kaul, K.L. Real-time PCR for detection and identification of Plasmodium spp. J. Clin. Microbiol. 2005, 43, 2435–2440. [Google Scholar] [CrossRef] [PubMed]
- Pholwat, S.; Liu, J.; Stroup, S.; Jacob, S.T.; Banura, P.; Moore, C.C.; Huang, F.; Laufer, M.K.; Houpt, E.; Guler, J.L. The Malaria TaqMan Array Card Includes 87 Assays for Plasmodium falciparum Drug Resistance, Identification of Species, and Genotyping in a Single Reaction. Antimicrob. Agents Chemother. 2017, 61, e00110-17. [Google Scholar] [CrossRef] [PubMed]
- Alemayehu, S.; Feghali, K.C.; Cowden, J.; Komisar, J.; Ockenhouse, C.F.; Kamau, E. Comparative evaluation of published real-time PCR assays for the detection of malaria following MIQE guidelines. Malar. J. 2013, 12, 277. [Google Scholar] [CrossRef] [PubMed]
- Craw, P.; Balachandran, W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip 2012, 12, 2469–2486. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Han, E.T.; Watanabe, R.; Sattabongkot, J.; Khuntirat, B.; Sirichaisinthop, J.; Iriko, H.; Jin, L.; Takeo, S.; Tsuboi, T. Detection of four Plasmodium species by genus and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 2007, 45, 2521–2528. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, D.P.; Savargaonkar, D.; Singh, O.P.; Bhatt, R.M.; Valecha, N. Evaluation of SYBR green I based visual loop-mediated isothermal amplification (LAMP) assay for genus and species-specific diagnosis of malaria in P. vivax and P. falciparum endemic regions. J. Vector Borne Dis. 2017, 54, 54–60. [Google Scholar] [PubMed]
- Serra-Casas, E.; Manrique, P.; Ding, X.C.; Carrasco-Escobar, G.; Alava, F.; Gave, A.; Rodriguez, H.; Contreras-Mancilla, J.; Rosas-Aguirre, A.; Speybroeck, N.; et al. Loop-mediated isothermal DNA amplification for asymptomatic malaria detection in challenging field settings: Technical performance and pilot implementation in the Peruvian Amazon. PLoS ONE 2017, 12, e0185742. [Google Scholar] [CrossRef] [PubMed]
- Marti, H.; Stalder, C.; González, I.J. Diagnostic accuracy of a LAMP kit for diagnosis of imported malaria in Switzerland. Travel Med. Infect. Dis. 2015, 13, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Lucchi, N.W.; Demas, A.; Narayanan, J.; Sumari, D.; Kabanywanyi, A.; Kachur, S.P.; Barnwell, J.W.; Udhayakumar, V. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS ONE 2010, 5, e13733. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Jimena, B.; Bekaert, M.; Bakheit, M.; Frischmann, S.; Patel, P.; Simon-Loriere, E.; Lambrechts, L.; Fall, C.; Faye, O.; Sall, A.A.; et al. Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype. PLoS Neglect. Trop. Dis. 2018. in submitted. [Google Scholar]
- Lopez-Jimena, B.; Wehner, S.; Harold, G.; Bakheit, M.; Frischmann, S.; Bekaert, M.; Faye, O.; Sall, A.A.; Weidmann, M. Development of a single-tube one-step RT-1 LAMP assay to detect the Chikungunya virus genome. PLoS Neglect. Trop. Dis. 2018. in submitted. [Google Scholar]
- Bass, C.; Nikou, D.; Vontas, J.; Donnelly, M.J.; Williamson, M.S.; Field, L.M. The Vector Population Monitoring Tool (VPMT): High-Throughput DNA-Based Diagnostics for the Monitoring of Mosquito Vector Populations. Malar. Res. Treat. 2010, 2010, 190434. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Williamson, M.S.; Field, L.M. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop. 2008, 107, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K.G.; Campbell-Lendrum, D.H.; Davies, C.R. A continental risk map for malaria mosquito (Diptera: Culicidae) vectors in Europe. J. Med. Entomol. 2002, 39, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Burkot, T.R.; Williams, J.L.; Schneider, I. Identification of Plasmodium falciparum-infected mosquitoes by a double antibody enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 1984, 33, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, R.A.; Burkot, T.R.; Andre, R.G.; Rosenberg, R.; Collins, W.E.; Roberts, D.R. Identification of Plasmodium vivax sporozoites in mosquitoes using an enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 1985, 34, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Fontenille, D.; Meunier, J.Y.; Nkondjio, C.A.; Tchuinkam, T. Use of circumsporozoite protein enzyme-linked immunosorbent assay compared with microscopic examination of salivary glands for calculation of malaria infectivity rates in mosquitoes (Diptera: Culicidae) from Cameroon. J. Med. Entomol. 2001, 38, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Durnez, L.; Van Bortel, W.; Denis, L.; Roelants, P.; Veracx, A.; Trung, H.D.; Sochantha, T.; Coosemans, M. False positive circumsporozoite protein ELISA: A challenge for the estimation of the entomological inoculation rate of malaria and for vector incrimination. Malar. J. 2011, 18, 195. [Google Scholar] [CrossRef] [PubMed]
- Beier, J.C.; Koros, J.K. Anatomical dissemination of circumsporozoite protein in wild Afrotropical Anopheles affects malaria sporozoite rate determination by ELISA. Med. Vet. Entomol. 1991, 5, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Snounou, G.; Singh, B. Nested PCR analysis of Plasmodium parasites. Methods Mol. Med. 2002, 72, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Boonsaeng, V.; Wilairat, P.; Panyim, S. Polymerase chain reaction detection of Plasmodium falciparum in mosquitoes. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 273–275. [Google Scholar] [CrossRef]
- Bass, C.; Nikou, D.; Blagborough, A.M.; Vontas, J.; Sinden, R.E.; Williamson, M.S.; Field, L.M. PCR-based detection of Plasmodium in Anopheles mosquitoes: A comparison of a new high-throughput assay with existing methods. Malar. J. 2008, 7, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kefi, M.; Mavridis, K.; Simões, M.L.; Dimopoulos, G.; Siden-Kiamos, I.; Vontas, J. New rapid one-step PCR diagnostic assay for Plasmodium falciparum infective mosquitoes. Sci. Rep. 2018. [CrossRef] [PubMed]
- Hemingway, J.; Field, L.; Vontas, J. An overview of insecticide resistance. Science 2002, 298, 96–97. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.J.; Isaacs, A.T.; Weetman, D. Identification, Validation, and Application of Molecular Diagnostics for Insecticide Resistance in Malaria Vectors. Trends Parasitol. 2016, 32, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Weetman, D.; Donnelly, M.J. Evolution of insecticide resistance diagnostics in malaria vectors. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C., IV; Vontas, J.G. Affordable assays for genotyping single nucleotide polymorphisms in insects. Insect Mol. Biol. 2007, 16, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Lüleyap, H.U.; Alptekin, D.; Kasap, H.; Kasap, M. Detection of knockdown resistance mutations in Anopheles sacharovi (Diptera: Culicidae) and genetic distance with Anopheles gambiae (Diptera: Culicidae) using cDNA sequencing of the voltage-gated sodium channel gene. J. Med. Entomol. 2002, 39, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Nikou, D.; Donnelly, M.J.; Williamson, M.S.; Ranson, H.; Ball, A.; Vontas, J.; Field, L.M. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: A comparison of two new high-throughput assays with existing methods. Malar. J. 2007, 6, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotakis, E.A.; Chaskopoulou, A.; Grigoraki, L.; Tsiamantas, A.; Kounadi, S.; Georgiou, L.; Vontas, J. Analysis of population structure and insecticide resistance in mosquitoes of the genus Culex, Anopheles and Aedes from different environments of Greece with a history of mosquito borne disease transmission. Acta Trop. 2017, 174, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.; Santos, J.M.; Martins, A.J. Mutations in the voltage-gated sodium channel gene of anophelines and their association with resistance to pyrethroids—A review. Parasite Vector 2014, 7, 450. [Google Scholar] [CrossRef] [PubMed]
- Chris, B.; Nikou, D.; Vontas, J.; Williamson, M.S.; Field, L.M. Development of high-throughput real-time PCR assays for the identification of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae. Pestic. Biochem. Phys. 2010, 96, 80–85. [Google Scholar] [CrossRef]
- Weill, M.; Malcolm, C.; Chandre, F.; Mogensen, K.; Berthomieu, A.; Marquine, M.; Raymond, M. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 2004, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nauen, R.; Vontas, J.; Kaussmann, M.; Wölfel, K. Pymetrozine is hydroxylated by CYP6CM1, a cytochrome P450 conferring neonicotinoid resistance in Bemisia tabaci. Pest Manag. Sci. 2013, 69, 457–461. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Strode, C.; Vontas, J.; Nikou, D.; Vaughan, A.; Pignatelli, P.M.; Louis, C.; Hemingway, J.; Ranson, H. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc. Natl. Acad. Sci. USA 2005, 102, 4080–4084. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Ismail, H.M.; Chandor-Proust, A.; Paine, M.J. Role of cytochrome P450s in insecticide resistance: Impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.; Ranson, H.; Alphey, L. Transcriptomics and disease vector control. BMC Biol. 2010, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.; Mitsakakis, K.; Zengerle, R.; Yewhalaw, D.; Sikaala, C.H.; Etang, J.; Fallani, M.; Carman, B.; Müller, P.; Chouaïbou, M.; et al. Automated innovative diagnostic, data management and communication tool, for improving malaria vector control in endemic settings. Studies in Health Technology and Informatics. Stud. Health Technol. Inform. 2016, 224, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Tropical infectious diseases: Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, S.; Tadesse, B.T.; Moussy, F.; Chua, A.; Zorzet, A.; Tangden, T.; Dolinger, D.L.; Page, A.L.; Crump, J.A.; D’Acremont, V.; et al. Target Product Profile for a Diagnostic Assay to Differentiate between Bacterial and Non-Bacterial Infections and Reduce Antimicrobial Overuse in Resource-Limited Settings: An Expert Consensus. PLoS ONE 2016, 11, e0161721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, N.P.; Vadnais, C.; Denkinger, C.; Engel, N.; Pai, M. Point-of-Care Testing for Infectious Diseases: Diversity, Complexity, and Barriers in Low- And Middle-Income Countries. PLoS Med. 2012, 9, e1001306. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, O.; Keller, M.; Schwemmer, F.; Zehnle, S.; Mark, D.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugal microfluidic platforms: Advanced unit operations and applications. Chem. Soc. Rev. 2015, 44, 6187–6229. [Google Scholar] [CrossRef] [PubMed]
- Rombach, M.; Kosse, D.; Faltin, B.; Wadle, S.; Roth, G.; Zengerle, R.; von Stetten, F. Real-time stability testing of air-dried primers and fluorogenic hydrolysis probes stabilized by trehalose and xanthan. Biotechniques 2014, 57, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Howson, E.L.A.; Armson, B.; Madi, M.; Kasanga, C.J.; Kandusi, S.; Sallu, R.; Chepkwony, E.; Siddle, A.; Martin, P.; Wood, J.; et al. Evaluation of Two Lyophilized Molecular Assays to Rapidly Detect Foot-and-Mouth Disease Virus Directly from Clinical Samples in Field Settings. Transbound. Emerg. Dis. 2017, 64, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-W.; Weissenberger, G.; Ching, W.-M. Development of Lyophilized Loop-Mediated Isothermal Amplification Reagents for the Detection of Leptospira. Mil. Med. 2016, 181, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [PubMed]
- Van Oordt, T.; Barb, Y.; Smetana, J.; Zengerle, R.; von Stetten, F. Miniature stick-packaging—An industrial technology for pre-storage and release of reagents in lab-on-a-chip systems. Lab Chip 2013, 13, 2888–2892. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, O.; Marquart, N.; Mark, D.; Roth, G.; Zengerle, R.; von Stetten, F. Real-time PCR based detection of a panel of food-borne pathogens on a centrifugal microfluidic “LabDisk” with on-disk quality controls and standards for quantification. Anal. Methods 2014, 6, 2038–2046. [Google Scholar] [CrossRef]
- Lole, K.S.; Arankalle, V.A. Quantitation of hepatitis B virus DNA by real-time PCR using internal amplification control and dual TaqMan MGB probes. J. Virol. Method 2006, 135, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Mitsakakis, K.; D’Acremont, V.; Strohmeier, O.; Hin, S.; Mark, D.; von Stetten, F.; Zengerle, R. Diagnostic tools and e-Health technologies for tackling febrile illness and enhancing patient management. Microelectron. Eng. 2018. in submitted. [Google Scholar]
- Tan, J.J.L.; Capozzoli, M.; Sato, M.; Watthanaworawit, W.; Ling, C.L.; Mauduit, M.; Malleret, B.; Gruner, A.C.; Tan, R.; Nosten, F.H.; et al. An integrated lab-on-chip for rapid identification and simultaneous differentiation of tropical pathogens. PLoS Neglect. Trop. Dis. 2014, 8, e3043. [Google Scholar] [CrossRef] [PubMed]
- Burn, J. Company profile: QuantuMDx group limited. Pharmacogenomics 2013, 14, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Czilwik, G.; Messinger, T.; Strohmeier, O.; Wadle, S.; von Stetten, F.; Paust, N.; Roth, G.; Zengerle, R.; Saarinen, P.; Niittymaki, J.; et al. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip 2015, 15, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, F.; Schwemmer, F.; Hutzenlaub, T.; Baumann, D.; Strohmeier, O.; Dingemanns, G.; Simons, G.; Sager, C.; Plobner, L.; von Stetten, F.; et al. LabDisk with complete reagent prestorage for sample-to-answer nucleic acid based detection of respiratory pathogens verified with influenza A H3N2 virus. Lab Chip 2016, 16, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Hin, S.; Lopez-Jimena, B.; Bakheit, M.; Klein, V.; Stack, S.; Fall, C.; Sall, A.; Enan, K.; Frischmann, S.; Gillies, L.; et al. The FeverDisk: Multiplex detection of fever-causing pathogens for rapid diagnosis of tropical diseases. In Proceedings of the 21st International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS 2017), Savannah, GA, USA, 22–26 October 2017; pp. 7–8. [Google Scholar]
- Africa’s Mobile Boom Powers Innovation Economy. Available online: http://www.bbc.com/news/business-28061813 (accessed on 3 November 2017).
- Brinkel, J.; Krämer, A.; Krumkamp, R.; May, J.; Fobil, J. Mobile Phone-Based mHealth Approaches for Public Health Surveillance in Sub-Saharan Africa: A Systematic Review. Int. J. Environ. Res. Public Health 2014, 11, 11559–11582. [Google Scholar] [CrossRef] [PubMed]
- Gove, S. Integrated management of childhood illness by outpatient health workers: Technical basis and overview. The WHO Working Group on Guidelines for Integrated Management of the Sick Child. Bull. World Health Organ. 1997, 75 (Suppl 1), 7–24. [Google Scholar] [PubMed]
- Mitchell, M.; Hedt-Gauthier, B.L.; Msellemu, D.; Nkaka, M.; Leshet, N. Using electronic technology to improve clinical care - results from a before-after cluster trial to evaluate assessment and classification of sick children according to Integrated Management of Childhood Illness (IMCI) protocol in Tanzania. BMC Med. Inform. Decis. 2013, 13, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaud-Althaus, C.; Shao, A.F.; Kahama-Maro, J.; Genton, B.; D’Acremont, V. Managing the Sick Child in the Era of Declining Malaria Transmission: Development of ALMANACH, an Electronic Algorithm for Appropriate Use of Antimicrobials. PLoS ONE 2015, 10, e0127674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, A.F.; Rambaud-Althaus, C.; Samaka, J.; Faustine, A.F.; Perri-Moore, S.; Swai, N.; Kahama-Maro, J.; Mitchell, M.; Genton, B.; D’Acremont, V. New Algorithm for Managing Childhood Illness Using Mobile Technology (ALMANACH): A Controlled Non-Inferiority Study on Clinical Outcome and Antibiotic Use in Tanzania. PLoS ONE 2015, 10, e0132316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keitel, K.; Kagoro, F.; Samaka, J.; Masimba, J.; Said, Z.; Temba, H.; Mlaganile, T.; Sangu, W.; Rambaud-Althaus, C.; Gervaix, A.; et al. A novel electronic algorithm using host biomarker point-of-care tests for the management of febrile illnesses in Tanzanian children (e-POCT): A randomized, controlled non-inferiority trial. PLoS Med. 2017, 14, e1002411. [Google Scholar] [CrossRef] [PubMed]
- Kasolo, F.; Yoti, Z.; Bakyaita, N.; Gaturuku, P.; Katz, R.; Fischer, J.E.; Perry, H.N. IDSR as a platform for implementing IHR in African countries. Biosecur. Bioterror. 2013, 11, 163–169. [Google Scholar] [CrossRef] [PubMed]
- District Health Information System 2. Available online: https://www.dhis2.org/ (accessed on 9 November 2017).
- Ohrt, C.; Roberts, K.W.; Sturrock, H.J.W.; Wegbreit, J.; Lee, B.Y.; Gosling, R.D. Information Systems to Support Surveillance for Malaria Elimination. Am. J. Trop. Med. Hyg. 2015, 93, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Fuentes, S.; Barker, C.M.; Coleman, M.; Coleman, M.; Park, B.; Reisen, W.K.; Eisen, L. Emerging Information Technologies to Provide Improved Decision Support for Surveillance, Prevention, and Control of Vector-Borne Diseases, Efficient Decision Support Systems-Practice and Challenges in Biomedical Related Domain; Jao, C., Ed.; InTech: London, UK, 2011. [Google Scholar] [CrossRef]
- Thomsen, E.K.; Deb, R.M.; Dunkley, S.; Coleman, M.; Foster, G.; Orlans, M.; Coleman, M. Enhancing Decision Support for Vector-Borne Disease Control Programs—The Disease Data Management System. PLoS Neglect. Trop. Dis. 2016, 10, e0004342. [Google Scholar] [CrossRef] [PubMed]
- Eisen, L.; Coleman, M.; Lozano-Fuentes, S.; McEachen, N.; Orlans, M.; Coleman, M. Multi-Disease Data Management System Platform for Vector-Borne Diseases. PLoS Neglect. Trop. Dis. 2011, 5, e1016. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, E.K.; Strode, C.; Hemmings, K.; Hughes, A.J.; Chanda, E.; Musapa, M.; Kamuliwo, M.; Phiri, F.N.; Muzia, L.; Chanda, J.; et al. Underpinning Sustainable Vector Control through Informed Insecticide Resistance Management. PLoS ONE 2014, 9, e99822. [Google Scholar] [CrossRef] [PubMed]
- Connolly, T.M.; Boyle, E.A.; MacArthur, E.; Hainey, T.; Boyle, J.M. A systematic literature review of empirical evidence on computer games and serious games. Comput. Educ. 2012, 59, 661–686. [Google Scholar] [CrossRef]
- Abt, C.C. Approach to methods of combined sociotechnological forecasting. Technol. Forecast Soc. Chang. 1970, 2, 17–22. [Google Scholar] [CrossRef]
- Luengo-Oroz, M.A.; Arranz, A.; Frean, J. Crowdsourcing Malaria Parasite Quantification: An Online Game for Analyzing Images of Infected Thick Blood Smears. J. Med. Internet Res. 2012, 14, 207–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavandadi, S.; Dimitrov, S.; Feng, S.; Yu, F.; Sikora, U.; Yaglidere, O.; Padmanabhan, S.; Nielsen, K.; Ozcan, A. Distributed Medical Image Analysis and Diagnosis through Crowd-Sourced Games: A Malaria Case Study. PLoS ONE 2012, 7, e37245. [Google Scholar] [CrossRef] [PubMed]
- Spotlight on Patent Pool. Available online: https://www.msfaccess.org/spotlight-on/patent-pool (accessed on 16 December 2017).
- Antinori, S.; Galimberti, L.; Milazzo, L.; Corbellino, M. Plasmodium knowlesi: The emerging zoonotic malaria parasite. Acta Trop. 2013, 125, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Lucchi, N.W.; Poorak, M.; Oberstaller, J.; DeBarry, J.; Srinivasamoorthy, G.; Goldman, I.; Xayavong, M.; da Silva, A.J.; Peterson, D.S.; Barnwell, J.W.; et al. A New Single-Step PCR Assay for the Detection of the Zoonotic Malaria Parasite Plasmodium knowlesi. PLoS ONE 2012, 7, e31848. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Kim Sung, L.; Matusop, A.; Radhakrishnan, A.; Shamsul, S.S.; Cox-Singh, J.; Thomas, A.; Conway, D.J. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 2004, 363, 1017–1024. [Google Scholar] [CrossRef]
- Franco, A.O.; Gomes, M.G.M.; Rowland, M.; Coleman, P.G.; Davies, C.R. Controlling Malaria Using Livestock-Based Interventions: A One Health Approach. PLoS ONE 2014, 9, e101699. [Google Scholar] [CrossRef] [PubMed]
- The malERA Refresh Consultative Panel on Tools for Malaria Elimination. malERA: An updated research agenda for diagnostics, drugs, vaccines, and vector control in malaria elimination and eradication. PLoS Med. 2017, 14, e1002455. [Google Scholar] [CrossRef]



| Technology Platform | Amplification Technology | Portability a | Degree of Multiplexity b | Time-to-Result | Stand-Alone c | Application in Malaria |
|---|---|---|---|---|---|---|
| GeneXpert® (Omni) | RT-qPCR | Benchtop (modular); Omni: portable | Up to 6 | 60 min | No; Omni: Yes | No |
| FilmArray® | Nested multiplex RT-qPCR; array-based detection | Benchtop | 27 | 60 min | No | No |
| AlereTM i | NEAR d (isothermal) | Portable | 2 | 15 min | Yes | No |
| Enigma® ML | RT-qPCR | Benchtop (modular) | 3 | 95 min | Yes | No |
| cobas® Liat | RT-qPCR | Portable | 3 | 20 min | Yes | No |
| EasyNATTM | CPA e (isothermal); visual readout in lateral flow strip | Handheld | 1 | 90 min | Yes | No |
| Verigene® RP Flex System | RT-qPCR, gold nanoparticle detection | Benchtop (modular) | 16 | 2 h | Yes | No |
| GenePOCTM | RT-qPCR | Benchtop | 12 | 60 min | Yes | No |
| Liaison® MDX | RT-qPCR | Benchtop | 4 | 60 min | No | No |
| VerePLEXTM Biosystem | PCR and microarray hybridization | Benchtop | 13 | ~3.5 h | No | In humans only |
| Q-POCTM | qPCR | Handheld | 5 | 15 min | Yes | In humans only |
| LabDisk (FeverDisk; demonstrator) | RT-LAMP | Portable | 12 | 2 h | No | In humans |
| LabDisk (VectorDisk; under development) | RT-qPCR | Portable | 36 | Target: 2 h | No | In vectors |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitsakakis, K.; Hin, S.; Müller, P.; Wipf, N.; Thomsen, E.; Coleman, M.; Zengerle, R.; Vontas, J.; Mavridis, K. Converging Human and Malaria Vector Diagnostics with Data Management towards an Integrated Holistic One Health Approach. Int. J. Environ. Res. Public Health 2018, 15, 259. https://doi.org/10.3390/ijerph15020259
Mitsakakis K, Hin S, Müller P, Wipf N, Thomsen E, Coleman M, Zengerle R, Vontas J, Mavridis K. Converging Human and Malaria Vector Diagnostics with Data Management towards an Integrated Holistic One Health Approach. International Journal of Environmental Research and Public Health. 2018; 15(2):259. https://doi.org/10.3390/ijerph15020259
Chicago/Turabian StyleMitsakakis, Konstantinos, Sebastian Hin, Pie Müller, Nadja Wipf, Edward Thomsen, Michael Coleman, Roland Zengerle, John Vontas, and Konstantinos Mavridis. 2018. "Converging Human and Malaria Vector Diagnostics with Data Management towards an Integrated Holistic One Health Approach" International Journal of Environmental Research and Public Health 15, no. 2: 259. https://doi.org/10.3390/ijerph15020259





