1. Introduction
Linear mixed models (LMM) have been used to find associations between continuous phenotypes and genetic variants, genes, and gene-environment (GE) interactions in unrelated and related subjects in genome-wide association (GWA) analysis. For unrelated subjects, the analysis can be performed within the generalized linear model framework, however, for related subjects as in the case of family data, one has to include the kinship matrix to take into account the correlation among the relatives for each family. In this paper, we are interested in testing GE interaction for discrete phenotypes. Generalized linear mixed models (GLMM) proposed by Breslow and Clayton [
1] is an ideal statistical approach to detect such an interaction with non-continuous phenotypes, because it can treat the familiar effect on the phenotype as a random effect.
Gene-based GE interaction tests have previously been proposed for independent subjects [
2,
3,
4]. While each GE interaction can be tested individually using one single nucleotide polymorphism (SNP) at a time, it is known that single SNP association is not as powerful as the gene-based analysis [
2] due to the linkage disequilibrium (LD) present among the SNPs in a gene. Lin et al. [
2] proposed a variance component test (VCT) of the interactions by treating the interactions as a random effect. This approach was extended to sequencing data with rare variants [
3,
5]. To overcome multicollinearity of the coefficients of the genetic markers, Lin et al. [
2,
3] applied ridge regression penalization of SNP coefficients and estimated the ridge penalty parameter with generalized cross-validation. However, this method is computationally demanding and their final test ignores the tuning of the ridge penalty parameter. Coombes [
6] instead proposed treating the genetic coefficients as a random effect in a linear mixed model framework to perform the ridge penalization. This equivalence was initially proposed by Bishop and Tipping [
7,
8] for Bayesian ridge regression in linear models framework. While this approach was able to incorporate the ridge penalty into the test statistic, it was only developed for a quantitative phenotype [
6].
Here, we propose a GLMM GE interaction framework for discrete and continuous phenotypes that treats the coefficients of genetic markers as random effects. Also, because the correlation among relatives cannot be ignored, this modeling framework incorporates the kinship matrix in the GLMM [
9]. We test for GE interaction between a set of SNPs and an environment by treating interaction coefficients as random effects using a VCT. Our proposed model includes three random effects: the first for genetic variants, the second for gene-environment interaction, and the third for the inclusion of families. In the methods section, we develop the framework for this model. We present the VCT as proposed by Lin [
10] in the presence of several random effects and adapt this VCT to accommodate the interactions [
11]. We also prove that the corresponding best linear unbiased predictor (BLUP) in our GLMM model is equivalent to the ridge regression estimator, as proposed by Shen et al. [
12]. In our simulations, we show that our model can be efficiently computed using the GMMAT package [
13] in
R and maintains appropriate type I error as well as sufficient power. Finally, we apply our methodology to test for genetic interactions with BMI associated with diabetes among the Baependi Heart Study [
14], which consists of 76 families of varying pedigree size.
3. Proposed GE Interaction Test
As mentioned by Lin et al. [
2], to treat
as a fixed vector and proceed with a
p degrees of freedom (DF) score test can result in loss of power to test for interaction. Another common strategy is to use a single SNP analysis of GE interaction, which assumes all SNPs are uncorrelated, however, this is usually not the case. In the majority of cases, the SNPs within a gene are highly correlated, thus, here we propose a test that accounts for correlation among SNPs and has uses less DF than the score test.
Using Equation (
4), our proposed test assumes
is a random vector following an arbitrary distribution with mean
and variance
. Thus, a test of the null hypothesis
is equivalent to testing
. For simplicity, we assume
.
In order to account for LD among SNPs in a gene and avoid estimation issues related to multicollinearity, we use a ridge regression approach to impose a penalty on
in the PQL proposed for GLMM [
1]. However, the selection of a penalty parameter can be computationally demanding [
2]. Thus, to expedite and incorporate the selection of a penalty parameter used in our proposed test, we specify
, as a random effect in Equation (
4), with
. By using this approach, we demonstrate later in
Section 3.1, under the null model, the best linear unbiased predictor (BLUP) of
is equivalent to the ridge regression estimator. Denoting
and
, and assuming
,
and
to be independent, we can write Equation (
4) in the GLMM form as
with
, where
is the
identity matrix and
, where
.
Based on the penalized quasi-likelihood (PQL) [
1], Lin [
10] developed a VCT for independent subjects in the framework of a GLMM. To test the null hypothesis
, the VCT uses the score statistic
where
and
are the maximum likelihood (ML) estimators for
and
, respectively, under the null model as described in
Section 3.1. In addition,
is the corresponding working vector, where
and
is calculated under the null model, where
denotes the derivate of function
. The covariance matrix for the working vector
is given by
.
Since
does not have a block diagonal structure, the score
cannot be written as a sum of
N independent random variables, corresponding to the families. Therefore, the asymptotic distribution of
is not a normal distribution, in contrast to the VCT of Lin [
10]. Instead, we follow the approach developed by Zhang and Lin [
11], and propose, as score statistic, the first term in Equation (
6), which corresponds to the quadratic form
To correct for bias, we use the restricted maximum likelihood (REML) estimators [
1] in the GLMM framework to obtain
and
under the null hypothesis.
Zhang and Lin [
11] showed that under
,
follows approximately a mixture of one degree of freedom, independent chi-square distributions. However, for computational ease, we use the Satterthwaite method [
15] to approximate the distribution of
by a scaled chi-square distribution
, where the scale parameter
and the degrees of freedom
can be calculated by equating the mean and variance of
to those of
.
When REML estimates are used to calculate
, Zhang and Lin [
11] showed that the mean and variance of
can be approximated, respectively, by
and
.
Dashed lines in and represent the cases: (i) known, implying a vector and matrix, respectively and (ii) unknown, implying a vector and matrix, respectively.
As the mean and variance of are given by and , respectively, we obtain the equations and , where denotes the matrix evaluated in . By solving these equations, we demonstrate that and .
Therefore, to test
, we propose the statistic
which follows approximately a chi-square distribution with
degrees of freedom.
3.1. Null Model Estimation
Our proposed score test requires that we first fit the null model. Under the null hypothesis
, Equation (
5) becomes
where
,
and
with
.
To estimate the parameters in Equation (
8), Breslow and Clayton [
1] proposed a Fisher scoring solution that may be expressed as the iterative solution to the system
This system is equivalent to the so called Henderson equations [
16] for computing the best linear unbiased estimator (BLUE) of
and the best linear unbiased predictor (BLUP) of
.
By re-expressing
to
in Equation (
9), we obtain the system
This new system is equivalent to using a ridge regression penalization for parameter
in Equation (
4) (see
Appendix A for details). Assuming that
is known, it can be shown that the solution of Equation (
10) is given by the following equations:
with
. Chen et al. [
17] fitted their GLMM by defining,
, and assuming
. However, the BLUP for
is identical to the sum of BLUPs in Equation (
11). Therefore, we use their PQL-based estimation algorithm implemented in the R package GMMAT (see Chen et al. [
17] and Chen and Conomos [
13] for details) to estimate our null model. The iterative process uses REML to estimate the variance parameters vector
used in the score statistic Equation (
7).
4. Simulations
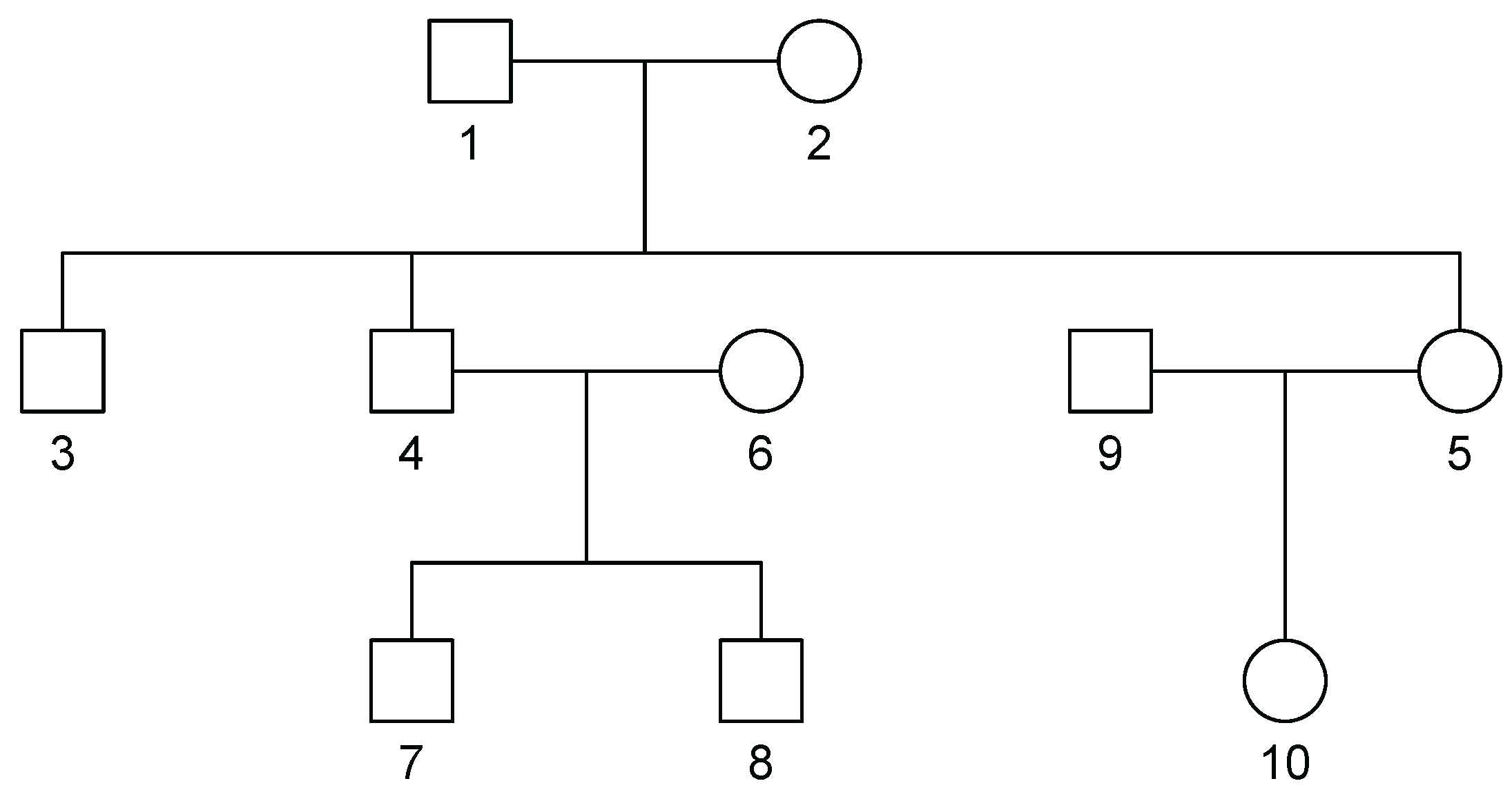
For our simulations, we used SimPed [
18] to generate 100 SNPs (50 independent and 50 in LD) for 1000 independent families with identical pedigree structures of size 10
Figure 1. For each simulation, we randomly sampled without replacement 100 families to obtain a sample of 1000 individuals.
The environment was simulated to be correlated within a family and depend on age and sex of the subject using the following model with parameters chosen to mimic our real data example:
where
is the indicator function,
where
is the
identity matrix, and
.
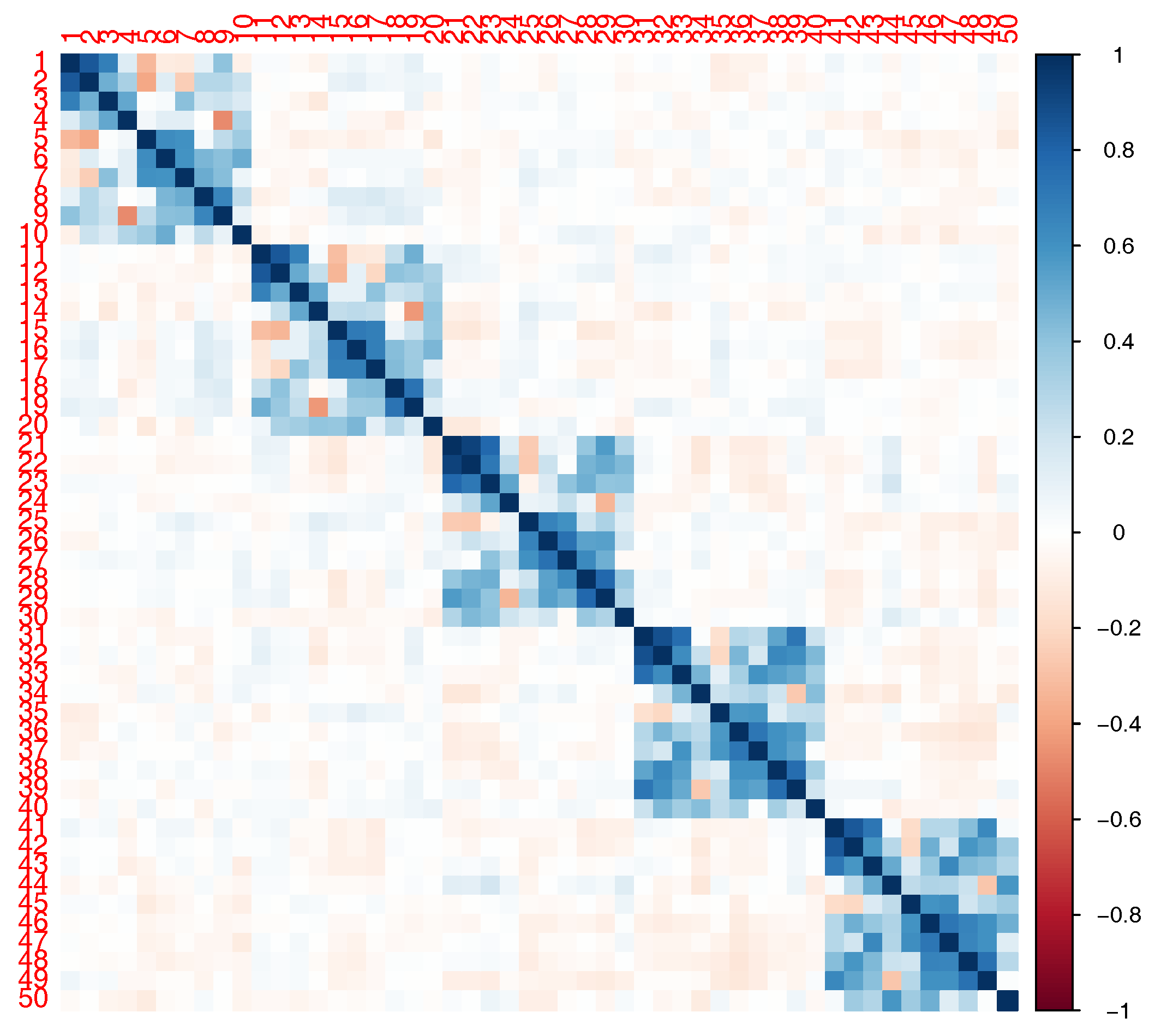
In our simulations, we considered a SNP-set with 50 independent SNPs or in LD. The correlation structure for the SNPs in LD is shown in
Figure 2. Using the
R package
SimCorMultRes [
19], we simulated a correlated binary phenotype dependent on family using the following mean structure:
with
, where
is the kinship matrix corresponding to the family pedigree in
Figure 1. Given
SimCorMultRes only allows for specification of the correlation matrix,
is set equal to 1.
and
are either independent SNPs (MAF = 0.3 and 0.1) or the first and fifth SNPs with MAF = 0.3 and 0.17 respectively from
Figure 2. Note that only the SNPs with a main effect interact with the environment in our model. We generated 10,000 and 1000 datasets to estimate type I error and empirical power, respectively, at an
level. Using these datasets, we compared the performances of the score test, MinP test, and our proposed VCT. As previously mentioned, the score test treats
as a fixed vector and results in a
p DF test. The null model for this test was estimated as specified in
Section 3.1. For the MinP test, which represents a single SNP analysis of GE interaction, we independently model the single SNP-by-environment interaction using Equation (
4) where
and
are a vector, rather than a matrix, for a SNP and GE interaction, respectively. For each model, we calculated the
p-value for the test of interaction and found the minimum
p-value among all tests. We corrected for multiple testing by multiplying by the number of effective SNPs in the gene [
20]. In the case of independence, the number of effective SNPs would be equivalent to the number of SNPs in the gene.
4.1. Type I Error
We first compared the empirical type I error of the different methods at 0.05
-level. To evaluate type I error, we set
and varied the number of SNPs
q in the gene. The SNPs were either independent or in LD. The empirical type I error rates are shown in
Table 1 as well as the mean of the fitted
,
, and
parameters from Equation (
8) across all simulations. While the variance component for the random effect defined by the kinship matrix stays approximately constant, the penalization term
increases as the number of SNPs in the model increases. Thus, like in ridge regression, increasing the number of parameters results in an increased penalization of the model. All of the methods were conservative in our simulations, but as
q increased, the score test became useless due to the large DF of the test.
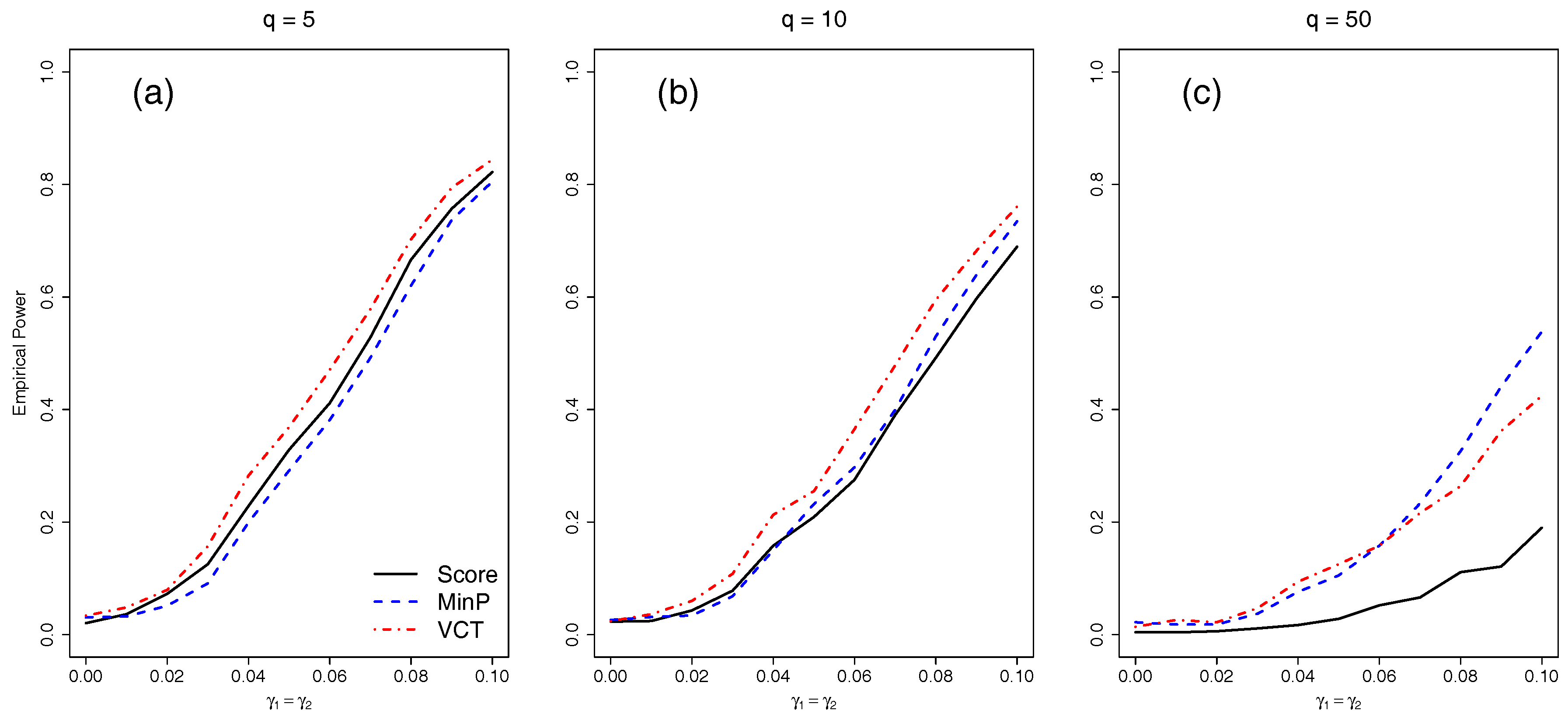
4.2. Empirical Power for Independent SNPs
We next compared the empirical power of the different methods with either five, 10, or 50 independent SNPs. We varied the amount of interaction for the two selected SNPs by varying
from 0 to 0.1 by 0.01.
Figure 3 shows that as the number of SNPs in the model increases, each of the methods lose power due to the increase in DF of each test. While the VCT performs best with five or 10 SNPs, the MinP test performs best for 50 SNPs because only two out of 50 SNPs have interaction. The MinP test will always perform best if very few of the SNPs in the set have interaction.
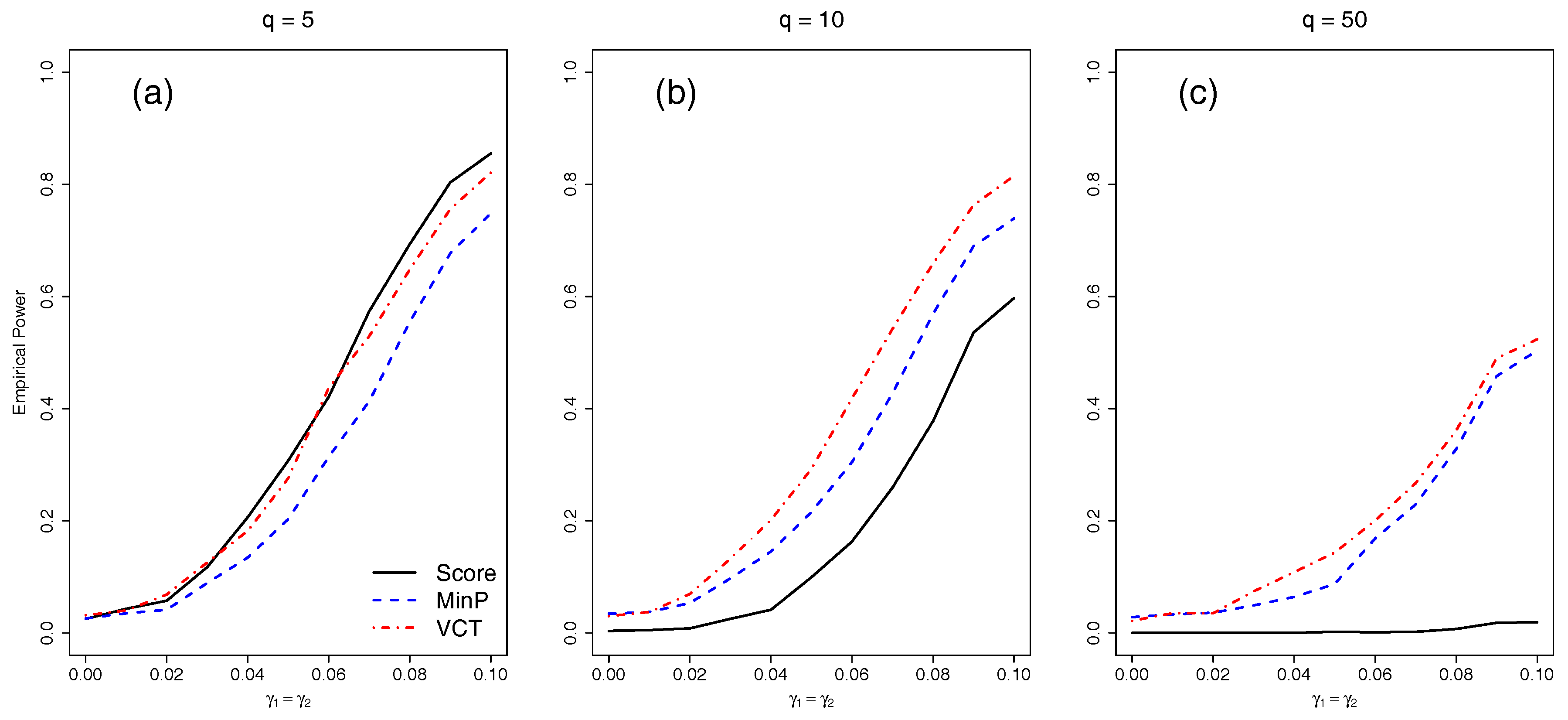
4.3. Empirical Power for SNPs in LD
Finally, we compared the empirical power of the different methods with either five, 10, or 50 SNPs in LD. We varied the amount of interaction for the two selected SNPs by varying
from 0 to 0.1 by 0.01.
Figure 4 shows that as before, each of the methods lose power as
q increases. The VCT outperforms the MinP test in all scenarios because the SNPs are correlated which the MinP test fails to account for.
5. Application to Baependi Data
We use our proposed method to test for GE interactions in the Baependi Heart Study [
14] between BMI and three different candidate genes that may be associated with type II diabetes (T2D). The first candidate gene we studied was the Peroxisome-Proliferator-Activated Receptors gamma (
PPARG) gene, which is a key regulator of adipocyte differentiation and energy balance. Two of the mutations in the
PPARG gene have been shown to be associated with obesity or diabetes-related phenotypes in different populations [
21].
PPARG2, the predominantly isoform of
PPARG, is expressed selectively and at a higher level in adipose tissue, where it modulates the expression of target genes implicated in adipocyte differentiation and glucose homeostasis [
22]. Thus, the
PPARG2 gene is a major candidate gene for T2D or obesity, both being complex phenotypes determined by the combination of multiple genetic and environmental factors [
23,
24]. The second candidate gene studied was the Fat Mass and Obesity associated protein (
FTO), which confers risk for obesity and BMI. Since obesity is known to be a predisposing factor for the development of T2D, it is not surprising that variants in FTO have been also found in T2D GWAS [
25]. The final candidate gene studied was the cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1 (
CDKAL1) gene which confers risk for obesity and T2D [
26]. In our study, the
PPARG,
FTO, and
CDKAL1 genes had 16, 149, and 186 genetic variants genotyped, respectively. However these numbers of SNPs do not represent the number of effective SNPs discussed in Gao et al. [
20], that is equivalent of the number of principal components to reach 99.5% of their total variation. Then, the effective number of SNPs associated with the
PPARG,
FTO, and
CDKAL1 genes are 10, 93, and 92, respectively. We used Equation (
5) to specify a logistic GLMM to test for GE interaction of the aforementioned genes with BMI associated with T2D status (case/control). Our model included age, sex, and the first two principal components of the entire genotype data of Baependi data as covariates. Due to some individuals missing genotype information for some SNPs, the tests of each gene had different sample sizes.
Table 2 describes the sample sizes (number of subjects and number of families) for cases and controls included in analysis of each gene.
We use our proposed method to test for GE interactions in the Baependi Heart Study [
14] between BMI and three different candidate genes that may be associated with type II diabetes (T2D). The first candidate gene we studied was the Peroxisome-Proliferator-Activated Receptors gamma (
PPARG) gene, which is a key regulator of adipocyte differentiation and energy balance. Two of the mutations in the
PPARG gene have been shown to be associated with obesity or diabetes-related phenotypes in different populations [
21].
PPARG2, the predominantly isoform of
PPARG, is expressed selectively and at a higher level in adipose tissue, where it modulates the expression of target genes implicated in adipocyte differentiation and glucose homeostasis [
22]. Thus, the
PPARG2 gene is a major candidate gene for T2D or obesity, both being complex phenotypes determined by the combination of multiple genetic and environmental factors [
23,
24]. The second candidate gene studied was the Fat Mass and Obesity associated protein (
FTO), which confers risk for obesity and BMI. Since obesity is known to be a predisposing factor for the development of T2D, it is not surprising that variants in FTO have been also found in T2D GWAS [
25]. The final candidate gene studied was the cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1 (
CDKAL1) gene which confers risk for obesity and T2D [
26]. In our study, the
PPARG,
FTO, and
CDKAL1 genes had 16, 149, and 186 genetic variants genotyped, respectively. However these numbers of SNPs do not represent the number of effective SNPs discussed in Gao et al. [
20], that is equivalent of the number of principal components to reach 99.5% of their total variation. Then, the effective number of SNPs associated with the
PPARG,
FTO, and
CDKAL1 genes are 10, 93, and 92, respectively. We used Equation (
5) to specify a logistic GLMM to test for GE interaction of the aforementioned genes with BMI associated with T2D status (case/control). Our model included age, sex, and the first two principal components of the entire genotype data of Baependi data as covariates. Due to some individuals missing genotype information for some SNPs, the tests of each gene had different sample sizes.
Table 2 describes the sample sizes (number of subjects and number of families) for cases and controls included in analysis of each gene.
In
Table 3, we report the
p-values for each method. By comparing the
p-values with respect to the corresponding
level, only the VCT identifies a significant GE interaction of BMI with
PPARG. All other tests were non-significant for this gene as well as for other candidate genes.
The variance estimates for families
and the ridge penalty
are reported in
Table 3. In
Section 4, we showed that the ridge penalty increases as the number of SNPs increases, however, our results for
PPARG,
FTO and
CDKAL1 suggest that
also depends on the number of subjects. Finally,
Table 3 also shows the execution times using the
R version 3.3.1 and a processor Intel(R) Core(TM) i5-6500 CPU @ 3.20 GHz with a RAM memory 8.00 GB and operating system 64-bits. Computation times for the VCT and the score test were considerably lower than those for the MinP test. The time to compute each test increased with the increase in number of SNPs in a gene and number of subjects in the analysis.