Correlation of CpG Island Methylation of the Cytochrome P450 2E1/2D6 Genes with Liver Injury Induced by Anti-Tuberculosis Drugs: A Nested Case-Control Study
Abstract
:1. Introduction
2. Materials and Method
2.1. Ethics Regulations
2.2. Sample Size Calculation
2.3. Selection of Experimental Cases and Controls
2.4. Epidemiological Investigation
2.5. Sample Collection and Extraction of Genomic DNA
2.6. Analysis of Gene Polymorphism
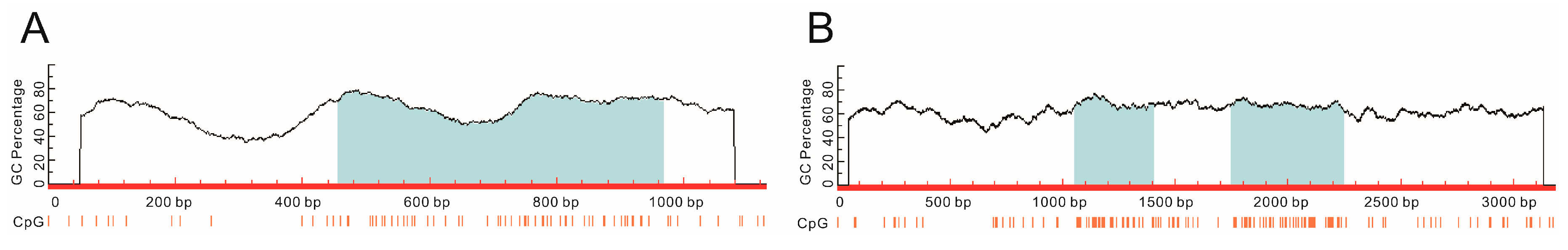
2.7. Methylation-Specific PCR Amplification
2.8. Statistical Analysis
3. Results
3.1. Basic Characteristics of the Cases
3.2. Correlation with Gene Polymorphism of the Two Drug-Metabolizing Enzymes and ADLI
3.3. Correlation between CpG Island Methylation of Genes of the Two Drug-Metabolizing Enzymes and ADLI
3.4. Multivariate Analysis of the Correlation between CpG Island Methylation of Genes of the Two Drug Metabolizing Enzymes and Anti-TB Drug-Induced Liver Injury
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schutz, C.; Ismail, Z.; Proxenos, C.J.; Marais, S.; Burton, R.; Kenyon, C.; Maartens, G.; Wilkinson, R.J.; Meintjes, G. Burden of antituberculosis and antiretroviral drug-induced liver injury at a secondary hospital in South Africa. S. Afr. Med. J. 2012, 102, 506–511. [Google Scholar] [PubMed]
- Yimer, G.; Ueda, N.; Habtewold, A.; Amogne, W.; Suda, A.; Riedel, K.D.; Burhenne, J.; Aderaye, G.; Lindquist, L.; Makonnen, E.; et al. Pharmacogenetic & pharmacokinetic biomarker for efavirenz based ARV and rifampicin based anti-TB drug induced liver injury in TB-HIV infected patients. PLoS ONE 2011, 6, e27810. [Google Scholar]
- Yee, D.; Valiquette, C.; Pelletier, M.; Parisien, I.; Rocher, I.; Menzies, D. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am. J. Respir. Crit. Care Med. 2003, 167, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Saeed, H.M.; Alanazi, M.S.; Nounou, H.A.; Salaby, M.A.; Semlali, A.; Azzam, N.; Aljebreen, A.; Alharby, O.; Parine, N.R.; Shaik, J.; et al. Cytochrome P450 1A1, 2E1 and GSTM1 gene polymorphisms and susceptibility to colorectal cancer in the Saudi population. Asian Pac. J. Cancer Prev. 2013, 14, 3761–3768. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Dinesh Kale, A.; Hallikerimath, S.; Yerramalla, V.; Subbiah, V.; Mishra, S. Association between GSTM1 and CYP1A1 polymorphisms and survival in oral cancer patients. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2013, 157, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Castillejos-López Mde, J.; García-Sancho, M.C.; Quiñones-Falconi, F.; Pérez-Padilla, J.R. Cytochrome P450 and NAT2 polymorphisms and drug metabolism in DOTS. Rev. Investig. Clin. 2008, 60, 47–57. [Google Scholar]
- Fukino, K.; Sasaki, Y.; Hirai, S.; Nakamura, T.; Hashimoto, M.; Yamagishi, F.; Ueno, K. Effects of N-acetyltransferase 2 (NAT2), CYP2E1 and glutathione-S-transferase (GST) genotypes on the serum concentrations of isoniazid and metabolites in tuberculosis patients. J. Toxicol. Sci. 2008, 33, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Chen, Y.; Xiang, Y.; Zhan, S. Drug-metabolising enzyme polymorphisms and predisposition to anti-tuberculosis drug-induced liver injury: A meta-analysis. Int. J. Tuberc. Lung Dis. 2008, 12, 994–1002. [Google Scholar] [PubMed]
- Turesky, R.J.; le Marchand, L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: Lessons learned from aromatic amines. Chem. Res. Toxicol. 2011, 24, 1169–1214. [Google Scholar] [CrossRef] [PubMed]
- Rendic, S.P.; Guengerich, F.P. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of chemicals. Chem. Res. Toxicol. 2015, 28, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Chern, H.D.; Su, W.J.; Wu, J.C.; Chang, S.C.; Chiang, C.H.; Chang, F.Y.; Lee, S.D. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology 2003, 37, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Liu, J.P.; Chowbay, B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab. Rev. 2009, 41, 89–295. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Kovalenko, V.M.; Bagnyukova, T.V.; Sergienko, O.V.; Bondarenko, L.B.; Shayakhmetova, G.M.; Matvienko, A.V.; Pogribny, I.P. Epigenetic changes in the rat livers induced by pyrazinamide treatment. Toxicol. Appl. Pharmacol. 2007, 225, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Sun, S.; Shen, L.; Zu, X.; Chen, Y.; Hao, J.; Huang, X.; Feng, F. DNA methylation in the rat livers induced by low dosage isoniazid treatment. Environ. Toxicol. Pharmacol. 2011, 32, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, B.; Sun, S.; Feng, F. Methylation of cytochrome p450 2E1 promoter induced by low dosage of isoniazid. Environ. Toxicol. Pharmacol. 2013, 36, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Schlesselman James, J.; Stolley Paul, D. Case-Control Studies: Design, Conduct, Analysis; Oxford University Press: New York, NY, USA, 1982; pp. 144–170. [Google Scholar]
- Tostmann, A.; Boeree, M.J.; Aarnoutse, R.E.; de Lange, W.C.; van der Ven, A.J.; Dekhuijzen, R. Antituberculosis drug-induced hepatotoxicity: Concise up-to-date review. J. Gastroenterol. Hepatol. 2008, 23, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Le Marchand, L.; Sivaraman, L.; Pierce, L.; Seifried, A.; Lum, A.; Wilkens, L.R.; Lau, A.F. Associations of CYP1A1, GSTM1, and CYP2E1 polymorphisms with lung cancer suggest cell type specificities to tobacco carcinogens. Cancer Res. 1998, 58, 4858–4863. [Google Scholar] [PubMed]
- Gao, Y.; Zhang, Q. Polymorphisms of the GSTM1 and CYP2D6 genes associated with susceptibility to lung cancer in Chinese. Mutat. Res. 1999, 444, 441–449. [Google Scholar] [CrossRef]
- Marez, D.; Legrand, M.; Sabbagh, N.; Lo Guidice, J.M.; Spire, C.; Lafitte, J.J.; Meyer, U.A.; Broly, F. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: Characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 1997, 7, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kang, H.S.; Jung, S.Y.; Min, S.Y.; Lee, S.; Kim, S.W.; Kwon, Y.; Lee, K.S.; Shin, K.H.; Ro, J. Methylation patterns of genes coding for drug-metabolizing enzymes in tamoxifen-resistant breast cancer tissues. J. Mol. Med. (Berl.) 2010, 88, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Sethi, G. Role of epigenetics in inflammation-associated diseases. Sub-Cell. Biochem. 2013, 61, 627–657. [Google Scholar]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zou, Z.Q.; Wang, L.Y.; Gao, S.; Fan, Y.C.; Long, B.; Xu, A.L.; Han, J.; Li, T.; Wang, K. Methylation of the glutathione-S-transferase M3 gene promoter is associated with oxidative stress in acute-on-chronic hepatitis B liver failure. Tohoku J. Exp. Med. 2012, 228, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Truong, P.K.; Lao, T.D.; Doan, T.P.; Le, T.A. BRCA1 promoter hypermethylation signature for early detection of breast cancer in the Vietnamese population. Asian Pac. J. Cancer Prev. 2014, 15, 9607–9610. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Bachman, K.E.; Herman, J.G.; Baylin, S.B. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001, 20, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Padjen, K.; Ratnam, S.; Storb, U. DNA methylation precedes chromatin modifications under the influence of the strain-specific modifier Ssm1. Mol. Cell. Biol. 2005, 25, 4782–4791. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Serra, L.; Esteller, M. Proteins that bind methylated DNA and human cancer: Reading the wrong words. Br. J. Cancer 2008, 98, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Circulating nucleic acids in plasma or serum. Clin. Chim. Acta 2001, 313, 143–146. [Google Scholar] [CrossRef]
- Vaissière, T.; Cuenin, C.; Paliwal, A.; Vineis, P.; Hoek, G.; Krzyzanowski, M.; Airoldi, L.; Dunning, A.; Garte, S.; Hainaut, P.; et al. Quantitative analysis of DNA methylation after whole bisulfitome amplification of a minute amount of DNA from body fluids. Epigenetics 2009, 4, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Yi, B.; Li, L.; Zhang, H.Y.; Sun, F.; Dong, S.Q.; Cao, Y. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp. Mol. Pathol. 2008, 85, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Snykers, S.; Henkens, T.; de Rop, E.; Vinken, M.; Fraczek, J.; de Kock, J.; De Prins, E.; Geerts, A.; Rogiers, V.; Vanhaecke, T. Role of epigenetics in liver-specific gene transcription, hepatocyte differentiation and stem cell reprogrammation. J. Hepatol. 2009, 51, 187–211. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Yu, E.; Shim, Y.H. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006, 233, 271–278. [Google Scholar] [CrossRef] [PubMed]
| Control | Case | Wald χ2 | p Value | OR (95% CI) | |
|---|---|---|---|---|---|
| CYP2E1 -1019 Mutant Genotype * | CYP2E1 -1019 Wild Genotype | ||||
| CYP2E1 -1019 Mutant genotype * | 10 | 24 | 0.667 | 0.414 | 0.694 (0.292–1.650) |
| CYP2E1 -1019 Wild genotype | 30 | 50 | |||
| CYP2E1 -1259 Mutant genotype * | CYP2E1 -1259 Wild genotype | ||||
| CYP2E1 -1259 Mutant genotype * | 16 | 18 | 2.174 | 0.140 | 1.651 (0.731–3.730) |
| CYP2E1 -1259 Wild genotype | 28 | 52 | |||
| CYP2D6 -188 Mutant genotype * | CYP2D6 -188 Wild genotype | ||||
| CYP2D6 -188 Mutant genotype * | 70 | 18 | 0.400 | 0.527 | 0.707 (0.216–2.312) |
| CYP2D6 -188 Wild genotype | 22 | 4 | |||
| CYP2D6 -4268 Mutant genotype * | CYP2D6 -4268 Wild genotype | ||||
| CYP2D6 -4268 Mutant genotype * | 92 | 16 | 6.150 | 0.013 | 0.250 (0.084–0.748) |
| CYP2D6 -4268 Wild genotype | 4 | 2 | |||
| Control | Case | Wald χ2 | p Value | OR (95% CI) | |
|---|---|---|---|---|---|
| CYP2E1 CpG Island Methylated * | CYP2E1 CpG Island Unmethylated | ||||
| CYP2E1 CpG island methylated * | 58 | 14 | 7.807 | 0.005 | 2.429 (1.303–4.525) |
| CYP2E1 CpG island unmethylated | 34 | 8 | |||
| CYP2D6 CpG island methylated * | CYP2D6 CpG island unmethylated | ||||
| CYP2D6 CpG island methylated * | 76 | 8 | 9.765 | 0.002 | 3.500 (1.595–7.679) |
| CYP2D6 CpG island unmethylated | 28 | 2 | |||
| Variables | S.E. | Wald χ2 | p Value | OR (95% CI) | |
|---|---|---|---|---|---|
| CYP2E1 CpG island methylated | 1.479 | 0.406 | 13.291 | <0.001 | 4.390 (1.982–9.724) |
| CYP2D6 CpG island methylated | 2.218 | 0.528 | 17.635 | <0.001 | 9.193 (3.264–25.888) |
| CYP2D6 -4268G/C mutant genotypes | −2.409 | 0.779 | 9.568 | 0.002 | 0.090 (0.020–0.414) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhu, X.; Li, Y.; Zhu, L.; Li, S.; Zheng, G.; Ren, Q.; Xiao, Y.; Feng, F. Correlation of CpG Island Methylation of the Cytochrome P450 2E1/2D6 Genes with Liver Injury Induced by Anti-Tuberculosis Drugs: A Nested Case-Control Study. Int. J. Environ. Res. Public Health 2016, 13, 776. https://doi.org/10.3390/ijerph13080776
Zhang J, Zhu X, Li Y, Zhu L, Li S, Zheng G, Ren Q, Xiao Y, Feng F. Correlation of CpG Island Methylation of the Cytochrome P450 2E1/2D6 Genes with Liver Injury Induced by Anti-Tuberculosis Drugs: A Nested Case-Control Study. International Journal of Environmental Research and Public Health. 2016; 13(8):776. https://doi.org/10.3390/ijerph13080776
Chicago/Turabian StyleZhang, Jinling, Xuebin Zhu, Yuhong Li, Lingyan Zhu, Shiming Li, Guoying Zheng, Qi Ren, Yonghong Xiao, and Fumin Feng. 2016. "Correlation of CpG Island Methylation of the Cytochrome P450 2E1/2D6 Genes with Liver Injury Induced by Anti-Tuberculosis Drugs: A Nested Case-Control Study" International Journal of Environmental Research and Public Health 13, no. 8: 776. https://doi.org/10.3390/ijerph13080776






