Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subject
2.2. Data Collection
2.3. Genotyping Analysis
2.4. Statistical Analysis
3. Results
3.1. Population Characteristics
3.2. Relationship between the Five Single Nucleotide Polymorphisms (SNPs) in GEMIN4 and AGO1 and Lung Cancer Risk
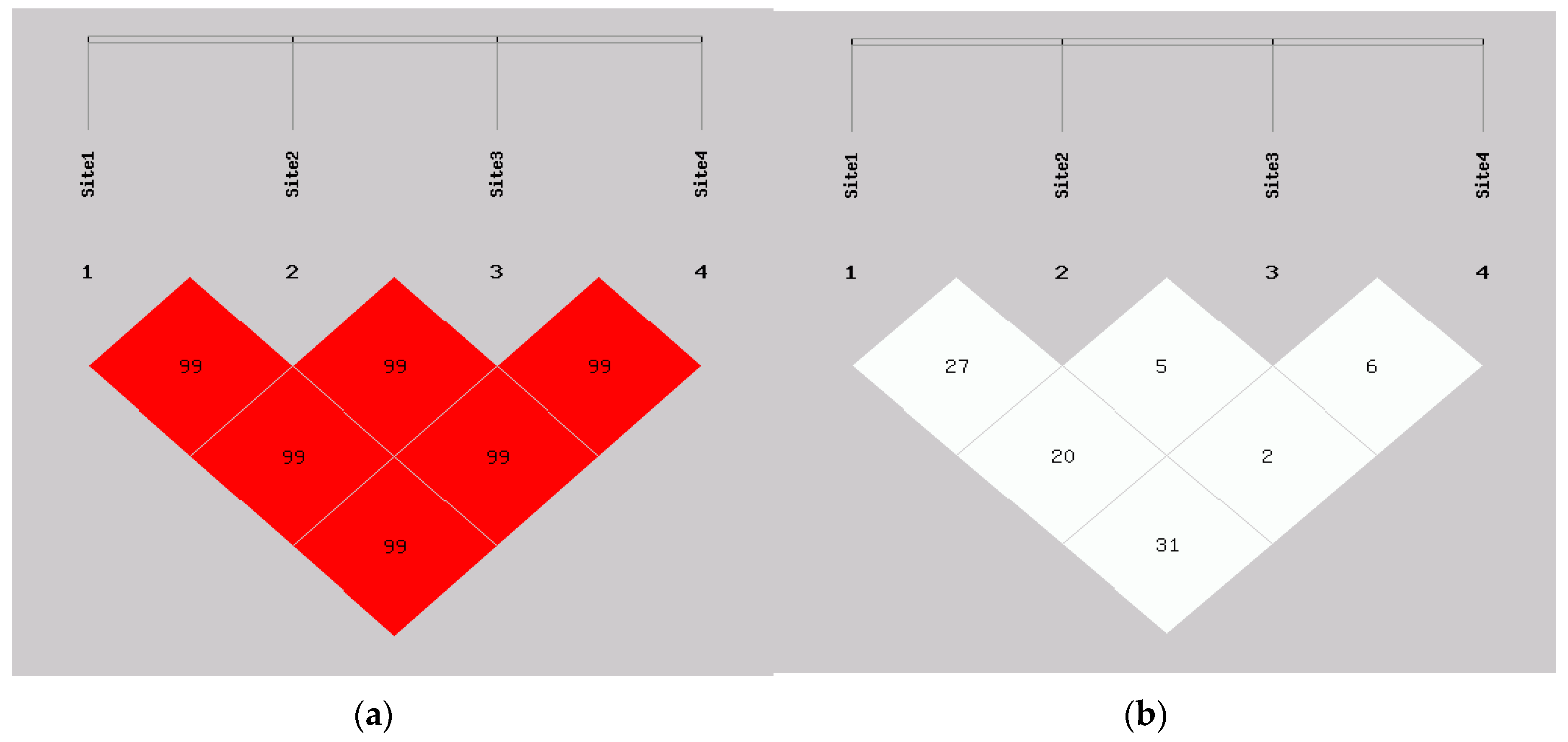
3.3. The Linkage Disequilibrium (LD) and Haplotype Analyses of the SNPs in GEMIN4 s and Lung Cancer Risk
3.4. Cumulative Effects of the Unfavorable Genotypes in Lung Adenocarcinoma
3.5. SNPs in GEMIN4 and AGO1 and Environmental Risk Factors (Cooking Oil Fume Exposure and Passive Smoking Exposure) as Well as Their Interaction on the Risk of Lung Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA A Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.K.; White, N.W.; Chan-Yeung, M.M. Lung cancer epidemiology and risk factors in Asia and Africa. Int. J. Tuberc. Lung Dis. 2004, 8, 1045–1057. [Google Scholar] [PubMed]
- Cortinovis, D.; Monica, V.; Pietrantonio, F.; Ceresoli, G.L.; La Spina, C.M.; Wannesson, L. Micrornas in non-small cell lung cancer: Current status and future therapeutic promises. Curr. Pharm. Des. 2014, 20, 3982–3990. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.L.; Chen, H.Y.; Chang, G.C.; Chen, C.Y.; Chen, H.W.; Singh, S.; Cheng, C.L.; Yu, C.J.; Lee, Y.C.; Chen, H.S.; et al. Microrna signature predicts survival and relapse in lung cancer. Cancer Cell 2008, 13, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.B.; Chen, J.P.; Tian, T.; Zhou, X.Y.; Gu, H.Y.; Xu, L.; Zeng, Y.; Miao, R.F.; Jin, G.F.; Ma, H.X.; et al. Genetic variants of mirna sequences and non-small cell lung cancer survival. J. Clin. Investig. 2008, 118, 2600–2608. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. Micrornas: Small rnas with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. Micrornas in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. Microrna signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small rnas in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Transcription and processing of human microrna precursors. Mol. Cell 2004, 16, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microrna networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. Trbp recruits the dicer complex to ago2 for microrna processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zhu, W.; Zhao, J.; He, J.; Qi, D.; Wang, L.; Ma, X.; Liu, P. Genetic variants in the microrna biosynthetic pathway gemin3 and gemin4 are associated with a risk of cancer: A meta-analysis. PeerJ 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Liu, J.N.; Wei, M.T.; He, Y.Z.; Liao, B.H.; Liao, G.; Li, H.; Huang, J. Genetic variants in the microrna machinery gene gemin4 are associated with risk of prostate cancer: A case-control study of the Chinese Han population. DNA Cell Biol. 2012, 31, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, Y.; Wood, C.G.; Yang, H.H.; Zhao, H.; Ye, Y.Q.; Gu, J.; Lin, J.; Habuchi, T.; Wu, X.F. Single nucleotide polymorphisms of microrna machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 2008, 14, 7956–7962. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Meyer, L.; Chang, D.W.; Lin, J.; Pu, X.; Ye, Y.Q.; Gu, J.A.; Wu, X.F.; Lu, K. Genetic variants in microrna biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010, 70, 9765–9776. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, J.; Wu, J.; Hu, Z.; Qin, Z.; Liu, X.; Guan, X.; Wang, Y.; Han, J.; Jiang, T.; et al. Evaluation of genetic variants in microrna biosynthesis genes and risk of breast cancer in Chinese women. Int. J. Cancer 2013, 133, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, Y.; Zhao, Y.; Guo, Z. Single-nucleotide polymorphisms of microrna processing machinery genes are associated with risk for gastric cancer. OncoTargets Ther. 2015, 8, 567–571. [Google Scholar]
- Li, M.C.; Yin, Z.H.; Guan, P.; Li, X.L.; Cui, Z.S.; Zhang, J.; Bai, W.J.; He, Q.C.; Zhou, B.S. Xrcc1 polymorphisms, cooking oil fume and lung cancer in Chinese women nonsmokers. Lung Cancer 2008, 62, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.H.; Li, H.; Cui, Z.G.; Ren, Y.W.; Li, X.L.; Wu, W.; Guan, P.; Qian, B.Y.; Rothman, N.; Lan, Q.; et al. Polymorphisms in pre-mirna genes and cooking oil fume exposure as well as their interaction on the risk of lung cancer in a chinese nonsmoking Female population. OncoTargets Ther. 2016, 9, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Hsiung, C.A.; Matsuo, K.; Hong, Y.C.; Seow, A.; Wang, Z.; Hosgood, H.D., 3rd; Chen, K.; Wang, J.C.; Chatterjee, N.; et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet. 2012, 44, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Zhang, Z.; He, Z.D.; Tang, W.; Li, T.; Zeng, Z.; He, L.; Shi, Y.Y. A partition-ligation-combination-subdivision em algorithm for haplotype inference with multiallelic markers: Update of the shesis (http://analysis. bio-x. cn). Cell Res. 2009, 19, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Alfredsson, L.; Kallberg, H.; Zdravkovic, S.; Ahlbom, A. Calculating measures of biological interaction. Eur. J. Epidemiol. 2005, 20, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Mendell, J.T. A small piece in the cancer puzzle: Micrornas as tumor suppressors and oncogenes. Oncogene 2006, 25, 6188–6196. [Google Scholar] [CrossRef] [PubMed]
- Lenkala, D.; Gamazon, E.R.; Lacroix, B.; Im, H.K.; Huang, R.S. Microrna biogenesis and cellular proliferation. Transl. Res. 2015, 166, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.; Mols, J.; Han, J.H. Risc-target interaction: Cleavage and translational suppression. Bba-Gene Regul. Mech. 2008, 1779, 668–677. [Google Scholar]
- Peters, L.; Meister, G. Argonaute proteins: Mediators of rna silencing. Mol. Cell 2007, 26, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mrna translation and stability by micrornas. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Qu, H.; Luo, M.; Wang, P.; Song, C.; Wang, K.; Zhang, J.; Dai, L. Micrornas related polymorphisms and genetic susceptibility to esophageal squamous cell carcinoma. Mol. Genet. Genom. 2014, 289, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Shi, Y.R.; Yin, Z.H.; Xue, X.X.; Zhou, B.S. An eight-mirna signature as a potential biomarker for predicting survival in lung adenocarcinoma. J. Transl. Med. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.S.; Dinney, C.P.; Ye, Y.Q.; Zhu, Y.; Grossman, H.B.; Wu, X.F. Evaluation of genetic variants in microrna-related genes and risk of bladder cancer. Cancer Res. 2008, 68, 2530–2537. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Choi, Y.Y.; Jin, G.; Kang, H.G.; Choi, J.E.; Jeon, H.S.; Lee, W.K.; Kim, D.S.; Kim, C.H.; Kim, Y.J.; et al. Association of a common ago1 variant with lung cancer risk: A two-stage case-control study. Mol. Carcin. 2010, 49, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, Y.B.N.; Jie, G.; Chi, Q.; Zhou, J.D.; Cui, B.B.; Piao, D.X.; Zhao, Y.S. Association between toll-like receptor 4 and interleukin 17 gene polymorphisms and colorectal cancer susceptibility in northeast China. Med. Oncol. 2014, 31, 73. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.H.; Cui, Z.G.; Guan, P.; Li, X.L.; Wu, W.; Ren, Y.W.; He, Q.C.; Zhou, B.S. Interaction between polymorphisms in pre-mirna genes and cooking oil fume exposure on the risk of lung cancer in Chinese non-smoking female population. PLoS ONE 2015, 10, e0128572. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.H.; Cui, Z.G.; Ren, Y.W.; Zhang, H.B.; Yan, Y.; Zhao, Y.X.; Ma, R.; Wang, Q.Q.; He, Q.C.; Zhou, B.S. Genetic polymorphisms of tert and clptm1l, cooking oil fume exposure, and risk of lung cancer: A case-control study in a Chinese non-smoking female population. Med. Oncol. 2014, 31, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, G.; Tan, W. Study on the effects of cooking oil fume condensate on the DNA integrity. J. Hyg. Res. 2002, 31, 238–240. [Google Scholar]
- Tokiwa, H.; Sera, N.; Nakashima, A.; Nakashima, K.; Nakanishi, Y.; Shigematu, N. Mutagenic and carcinogenic significance and the possible induction of lung cancer by nitro aromatic hydrocarbons in particulate pollutants. Environ. Health Perspect. 1994, 102 (Suppl. 4), 107–110. [Google Scholar] [CrossRef]
| Variables | Cases (%) | Controls (%) | p Value |
|---|---|---|---|
| Females | 473 | 395 | |
| Mean age (years) | 56.26 ± 11.71 | 56.13 ± 11.64 | 0.87 |
| Histological | |||
| Adenocarcinoma | 321 (67.9%) | ||
| Squamous cell carcinoma | 65 (13.7%) | ||
| SCLC | 66 (14.0%) | ||
| Others a | 21 (4.4%) |
| Chr Location | Gene | SNP | Position | Major/Minor Allele |
|---|---|---|---|---|
| 17p13 | GEMIN4 | rs7813 | C1022R | T/C |
| 17p13 | GEMIN4 | rs2740349 | N918D | A/G |
| 17p13 | GEMIN4 | rs2291778 | Intron | G/T |
| 17p13 | GEMIN4 | rs910924 | Promoter | C/T |
| 1p34.3 | AGO1(EIF2C1) | rs595961 | Intron | G/A |
| SNP | Genotype | Lung Cancer Cases (%) N = 473 | Controls (%) N = 395 | p of HWE | Adjusted OR a | 95% CI | p |
|---|---|---|---|---|---|---|---|
| rs7813 | TT | 242 (51.2) | 153 (38.7) | 0.320 | Ref | ||
| CT | 177 (37.4) | 193 (48.9) | 0.580 | 0.435, 0.773 | <0.001 * | ||
| CC | 54 (11.4) | 49 (12.4) | 0.694 | 0.448, 1.075 | 0.102 | ||
| Dominant model | CT + CC | 231 (48.8) | 242 (61.3) | 0.604 | 0.460, 0.792 | <0.001 * | |
| Additive model | C allele | 0.740 | 0.605, 0.904 | 0.003 * | |||
| rs2740349 | AA | 375 (79.3) | 298 (75.4) | 0.123 | Ref | ||
| AG | 93 (19.7) | 86 (21.8) | 0.859 | 0.617, 1.195 | 0.367 | ||
| GG | 5 (1.1) | 11 (2.8) | 0.361 | 0.124, 1.051 | 0.062 | ||
| Dominant model | AG + GG | 98 (20.7) | 97 (24.6) | 0.803 | 0.584, 1.106 | 0.179 | |
| Additive mode | G allele | 0.772 | 0.579, 1.030 | 0.078 | |||
| rs2291778 | GG | 225 (47.6) | 214 (54.2) | 0.513 | Ref | ||
| GT | 196 (41.4) | 150 (38.0) | 1.239 | 0.933, 1.645 | 0.139 | ||
| TT | 52 (11.0) | 31 (7.8) | 1.596 | 0.983, 2.591 | 0.059 | ||
| Dominant model | GT + TT | 248 (52.4) | 181 (45.8) | 1.303 | 0.996, 1.704 | 0.053 | |
| Additive mode | T allele | 1.265 | 1.027, 1.559 | 0.027 * | |||
| rs910924 | CC | 369 (78.0) | 277 (70.1) | 0.891 | Ref | ||
| CT | 96 (20.3) | 108 (27.3) | 0.667 | 0.487, 0.915 | 0.012 * | ||
| TT | 8 (1.7) | 10 (2.5) | 0.600 | 0.234, 1.541 | 0.289 | ||
| Dominant model | CT + TT | 104 (22.0) | 118 (29.9) | 0.662 | 0.487, 0.899 | 0.008 * | |
| Additive mode | T allele | 0.695 | 0.528, 0.913 | 0.009 * | |||
| rs595961 | GG | 293 (61.9) | 285 (72.2) | 0.748 | Ref | ||
| AG | 167 (35.3) | 102 (25.8) | 1.593 | 1.186, 2.141 | 0.002 * | ||
| AA | 13 (2.7) | 8 (2.0) | 1.580 | 0.645, 3.871 | 0.317 | ||
| Dominant model | AG + AA | 180 (38.1) | 110 (27.8) | 1.592 | 1.194, 2.123 | 0.002 * | |
| Additive mode | A allele | 1.460 | 1.135, 1.878 | 0.003 * |
| SNP | Genotype | Controls (%) | Adenocarcinoma (%) | Adjusted OR a | 95% CI | p |
|---|---|---|---|---|---|---|
| N = 395 | N = 321 | |||||
| rs7813 | TT | 153 (38.7) | 170 (53.0) | Ref | ||
| CT | 193 (48.9) | 118 (36.8) | 0.550 | 0.401, 0.755 | <0.001 * | |
| CC | 49 (12.4) | 33 (10.3) | 0.595 | 0.362, 0.976 | 0.040 * | |
| Dominant model | CT + CC | 242 (61.3) | 151 (47.0) | 0.562 | 0.417, 0.757 | <0.001 * |
| Additive model | C allele | 0.689 | 0.551, 0.862 | 0.001 * | ||
| rs2740349 | AA | 298 (75.4) | 259 (80.7) | Ref | ||
| AG | 86 (21.8) | 59 (18.4) | 0.788 | 0.543, 1.141 | 0.207 | |
| GG | 11 (2.8) | 3 (0.9) | 0.313 | 0.086, 1.134 | 0.077 | |
| Dominant model | AG + GG | 97 (24.6) | 62 (19.3) | 0.734 | 0.512, 1.052 | 0.093 |
| Additive model | G allele | 0.711 | 0.513, 0.987 | 0.041 * | ||
| rs2291778 | GG | 214 (54.2) | 150 (46.7) | Ref | ||
| GT | 150 (38.0) | 134 (41.7) | 1.275 | 0.932, 1.745 | 0.128 | |
| TT | 31 (7.8) | 37 (11.5) | 1.719 | 1.017, 2.904 | 0.043 * | |
| Dominant model | GT + TT | 181 (45.8) | 171 (53.3) | 1.354 | 1.006, 1.821 | 0.045 * |
| Additive model | T allele | 1.307 | 1.040, 1.642 | 0.021 * | ||
| rs910924 | CC | 277 (70.1) | 261 (81.3) | Ref | ||
| CT | 108 (27.3) | 55 (17.1) | 0.540 | 0.374, 0.779 | 0.001 * | |
| TT | 10 (2.5) | 5 (1.6) | 0.533 | 0.180, 1.582 | 0.257 | |
| Dominant model | CT + TT | 118 (29.9) | 60 (18.7) | 0.540 | 0.379, 0.769 | 0.001 * |
| Additive model | T allele | 0.583 | 0.424, 0.802 | 0.001 * | ||
| rs595961 | GG | 285 (72.2) | 198 (61.7) | Ref | ||
| AG | 102 (25.8) | 112 (34.9) | 1.580 | 1.143, 2.184 | 0.006 * | |
| AA | 8 (2.0) | 11 (3.4) | 1.982 | 0.783, 5.018 | 0.149 | |
| Dominant model | AG + AA | 110 (27.8) | 123 (38.3) | 1.609 | 1.175, 2.205 | 0.003 * |
| Additive model | A allele | 1.502 | 1.143, 1.974 | 0.003 * |
| Haplotype a | Controls (%) | Lung Cancer | Adenocarcinoma | ||||
|---|---|---|---|---|---|---|---|
| N (%) | OR (95% CI) | p | N (%) | OR (95% CI) | p | ||
| TAGC | 287 (36.3) | 360 (38.1) | 1.080 (0.889, 1.314) | 0.438 | 250 (38.9) | 1.118 (0.901, 1.386) | 0.310 |
| TATC | 212 (26.8) | 300 (31.7) | 1.265 (1.027, 1.559) | 0.027 * | 208 (32.4) | 1.307 (1.040, 1.642) | 0.021 * |
| CAGT | 128 (16.2) | 111 (11.7) | 0.688 (0.523, 0.905) | 0.007 * | 65 (10.1) | 0.583 (0.424, 0.801) | 0.001 * |
| CGGC | 108 (13.7) | 103 (10.9) | 0.772 (0.579, 1.030) | 0.078 | 65 (10.1) | 0.711 (0.513, 0.986) | 0.040 * |
| CAGC | 55 (7) | 71 (7.5) | 1.087 (0.754, 1.566) | 0.655 | 54 (8.4) | 1.227 (0.830, 1.814) | 0.303 |
| Number of Unfavorable Genotypes a | Adenocarcinoma (%) | Controls (%) | Adjusted OR b | 95% CI | p |
|---|---|---|---|---|---|
| 0/1 | 41 (12.8) | 100 (25.3) | Ref | ||
| 2/3 | 137 (42.7) | 186 (47.1) | 1.798 | 1.175, 2.751 | 0.007 * |
| 4/5 | 143 (44.5) | 109 (27.6) | 3.206 | 2.063, 4.983 | <0.001 * |
| Cooking Oil Fume Exposure | Genotype | Cases (%) | Controls (%) | Adjusted OR a | 95% CI | p Value | Passive Smoking Exposure | Cases (%) | Controls (%) | Adjusted OR a | 95% CI | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7813 | ||||||||||||
| − | CT + CC | 68 (30.4) | 118 (48.4) | Ref | − | 46 (20.5) | 62 (25.4) | Ref | ||||
| − | TT | 74 (33.0) | 74 (30.3) | 1.836 | 1.073, 3.143 | 0.027 * | − | 47 (21.0) | 54 (22.1) | 1.189 | 0.613, 2.305 | 0.609 |
| + | CT + CC | 46 (20.5) | 36 (14.8) | 2.266 | 1.182, 4.343 | 0.014 * | + | 68 (30.4) | 92 (37.7) | 1.022 | 0.564, 1.851 | 0.940 |
| + | TT | 36 (16.1) | 16 (6.6) | 1.778 | 0.688, 4.598 | 0.235 | + | 63 (28.1) | 36 (14.8) | 2.578 | 1.266, 5.252 | 0.009 * |
| rs2740349 | ||||||||||||
| − | AG + GG | 26 (11.6) | 50 (20.5) | Ref | − | 19 (8.5) | 34 (13.9) | Ref | ||||
| − | AA | 116 (51.8) | 142 (58.2) | 1.746 | 0.920, 3.313 | 0.088 | − | 74 (33.0) | 82 (33.6) | 1.586 | 0.739, 3.402 | 0.237 |
| + | AG + GG | 18 (8.0) | 14 (5.7) | 2.384 | 0.834, 6.808 | 0.105 | + | 25 (11.2) | 30 (12.3) | 1.493 | 0.596, 3.741 | 0.392 |
| + | AA | 64 (28.6) | 38 (15.6) | 1.316 | 0.472, 3.669 | 0.600 | + | 106 (47.3) | 98 (40.2) | 2.065 | 0.974, 4.379 | 0.059 |
| rs2291778 | ||||||||||||
| − | GG | 64 (28.6) | 84 (34.4) | Ref | − | 45 (20.1) | 54 (22.1) | Ref | ||||
| − | GT + TT | 78 (34.8) | 108 (44.3) | 0.992 | 0.583, 1.685 | 0.975 | − | 48 (21.4) | 62 (25.4) | 0.903 | 0.465, 1.752 | 0.762 |
| + | GG | 40 (17.9) | 26 (10.7) | 2.028 | 0.969, 4.245 | 0.061 | + | 59 (26.3) | 56 (23.0) | 1.263 | 0.655, 2.437 | 0.486 |
| + | GT + TT | 42 (18.8) | 26 (10.7) | 2.157 | 1.031, 4.512 | 0.041 * | + | 72 (32.1) | 72 (29.5) | 1.325 | 0.700, 2.511 | 0.388 |
| rs910924 | ||||||||||||
| − | CT + TT | 32 (14.3) | 56 (23.0) | Ref | − | 20 (8.9) | 30 (12.3) | Ref | ||||
| − | CC | 110 (49.1) | 136 (55.7) | 1.437 | 0.788, 2.623 | 0.237 | − | 73 (32.6) | 86 (35.2) | 1.296 | 0.597, 2.814 | 0.512 |
| + | CT + TT | 19 (8.5) | 18 (7.4) | 1.860 | 0.721, 4.799 | 0.199 | + | 31 (13.8) | 44 (18.0) | 1.126 | 0.468, 2.707 | 0.792 |
| + | CC | 63 (28.1) | 34 (13.9) | 1.755 | 0.674, 4.572 | 0.249 | + | 100 (44.6) | 84 (34.4) | 1.863 | 0.864, 4.016 | 0.112 |
| Rs595961 | ||||||||||||
| − | GG | 93 (41.5) | 144 (59.0) | Ref | − | 56 (25.0) | 84 (34.4) | Ref | ||||
| − | AG + AA | 49 (21.9) | 48 (19.7) | 1.594 | 0.982, 2.587 | 0.059 | − | 37 (16.5) | 32 (13.1) | 0.978 | 0.955, 1.002 | 0.072 |
| + | GG | 50 (22.3) | 44 (18.0) | 1.726 | 1.060, 2.810 | 0.028 * | + | 87 (38.8) | 104 (42.6) | 1.333 | 0.850, 2.091 | 0.210 |
| + | AG + AA | 32 (14.3) | 8 (3.3) | 6.314 | 2.752, 14.485 | <0.001 * | + | 44(19.6) | 24 (9.8) | 3.139 | 1.678, 5.871 | <0.001 * |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Yin, Z.; Li, X.; Xia, L.; Zhou, B. Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers. Int. J. Environ. Res. Public Health 2016, 13, 939. https://doi.org/10.3390/ijerph13100939
Fang X, Yin Z, Li X, Xia L, Zhou B. Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers. International Journal of Environmental Research and Public Health. 2016; 13(10):939. https://doi.org/10.3390/ijerph13100939
Chicago/Turabian StyleFang, Xue, Zhihua Yin, Xuelian Li, Lingzi Xia, and Baosen Zhou. 2016. "Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers" International Journal of Environmental Research and Public Health 13, no. 10: 939. https://doi.org/10.3390/ijerph13100939






