Removal of Fecal Indicators, Pathogenic Bacteria, Adenovirus, Cryptosporidium and Giardia (oo)cysts in Waste Stabilization Ponds in Northern and Eastern Australia
Abstract
:1. Introduction
2. Experimental Section
2.1. Sampling Sites
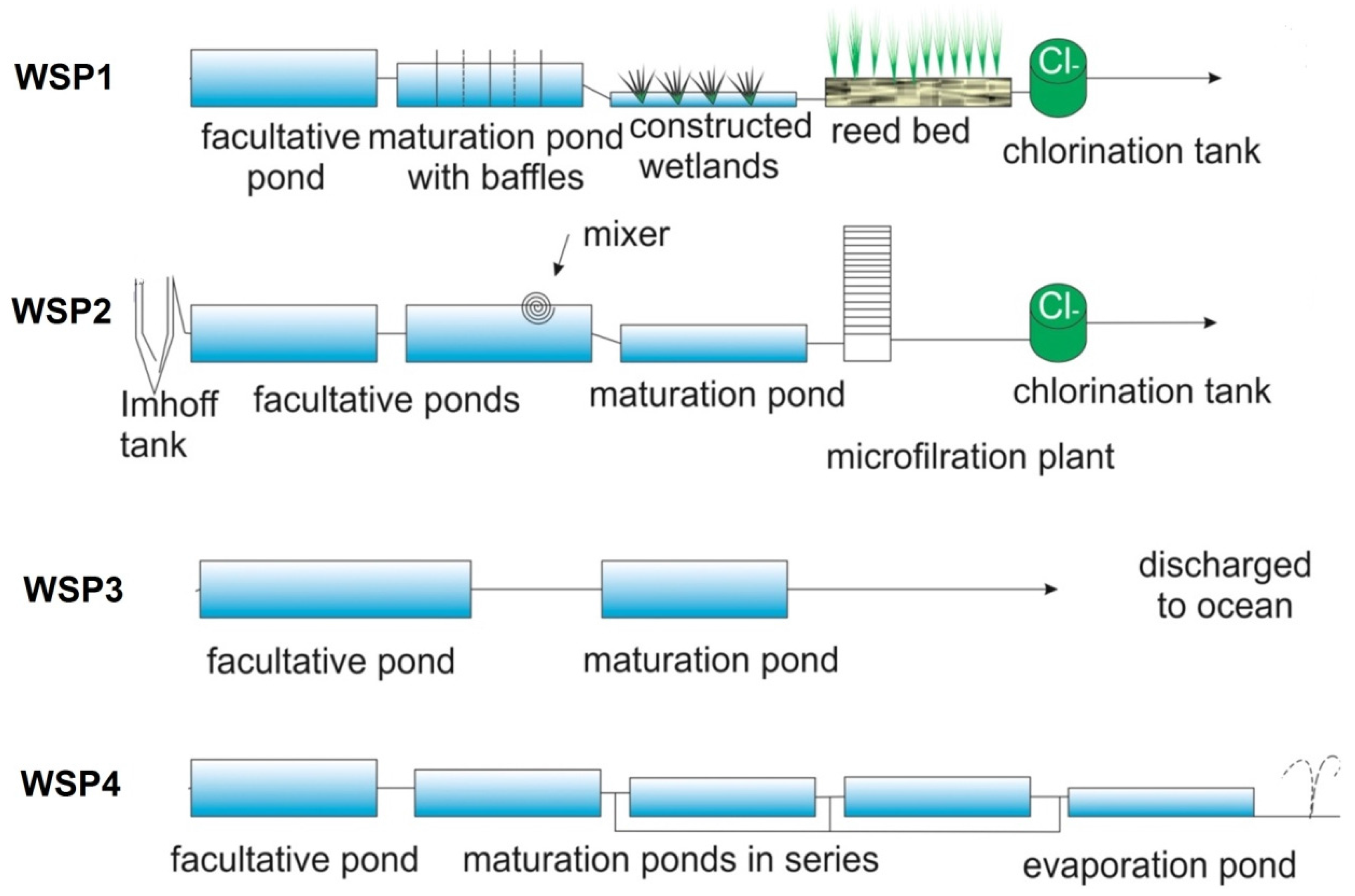
2.2. Physico-Chemical Measurements
| Ponds | Zone | Year | T °C | pH | DO, mg/L | Turbidity, NTU | Conductivity, µS/cm |
|---|---|---|---|---|---|---|---|
| WSP1 | subtropical | 2014 | 17–22 | 8.8–9.4 | 8–23.7 | 26–72 | 842–1041 |
| WSP2 | subtropical | 2014 | 13.6–17.5 | 10.2–10.6 | 17.4–24.4 | 34–66 | 929–1002 |
| WSP3 | tropical | 2013 | 28.7–38.0 | 6.08–10.45 | 0–34.26 | 76–136 | 467–766 |
| WSP4 | tropical | 2014 | 25–26.4 | 7.8–9.2 | 9.9–11.4 | 27–65 | 1750–1876 |
2.3. Culture Based Bacterial Quantification
2.4. F+ RNA Coliphage Quantification
2.5. Viral Precipitation and Infectivity Assay
2.6. Cryptosporidium and Giardia Enumeration
2.7. DNA Extraction and Recovery
2.8. Preparation of Stocks for qPCR Calibration Curves
2.9. PCR Primers and TaqMan Probes
2.10. Real-Time qPCR
| Microorganisms | Gene | Function | Sequences (5′-3′) | Primer Concentration, µM | Reference |
|---|---|---|---|---|---|
| E. coli | uidA | glucuronidase | F: GTGTGATATCTACCCGCTTCGC | 0.7 | [14] |
| R: AGAACGGTTTGTGGTTAATCAGGA | 0.7 | ||||
| P: TCGGCATCCGGTCAGTGGCAGT | 0.2 | ||||
| Enterococcus faecalis | 23s RNA | Ribosomal gene | F: GAGAAATTCCAAACGAACTTG | 0.5 | [15] |
| R: CAGTGCTCTACCTCCATCATT | 0.5 | ||||
| P: TGGTTCTCTCCGAAATAGCTTTAGGGCTA | 0.08 | ||||
| DNA IAC pM13mp18 | NA | F: AAGATTTGAATCGGTTGCTTGG | 0.4 | [13] | |
| R: GCCACTGCTCCATTATCTGG | 0.4 | ||||
| P: CCGATTGTTAGCCAGCCCATGCCA | 0.2 | ||||
| Adenovirus (all types) | Hexon gene | F: GCCACGGTGGGGTTTCTAAACTT | 0.5 | [16] | |
| R: GCCCCAGTGGTCTTACATGCA | 0.5 | ||||
| P: TGCACCAGACCCGGGCTCAGGAGGTACTCCGA | 0.2 | ||||
| Campylobacter jejuni | mapA | Mucus adhesion-promoting protein | F: GGTTTTGAAGCAAAGATTAAAGG | 0.5 | [17] |
| R: AAGCAATACCAGTGTCTAAAGTGC | 0.5 | [17] | |||
| P: TGGCACAACATTGAATTCCAACATCGCTA | 0.3 | [18] | |||
| VS1 | n/a | F: GAATGAAATTTTAGAATGGGG | 0.4 | [19] | |
| R: GATATGTATGATTTTATCCTGC | 0.4 | ||||
| P: TTTAACTTGGCTAAAGGCTAAGGCT | 0.1 | ||||
| Salmonella enterica | gyrB | gyrase protein | F: CGTGGGCGTCTCGGTAGTY | 0.5 | [20] |
| R: CTCATATTCAAATTCAGTGACG | 0.5 | [20] | |||
| P: AAACCGGCACGATGGTACGTTTCT | 0.25 | This study | |||
| ttrRSBCA | tetrathionate respiration | F: CTCACCAGGAGATTACAACATGG | 0.4 | [21] | |
| R: AGCTCAGACCAAAAGTGACCATC | 0.4 | ||||
| P: CG +ACG +GCG +AG+ACCG | 0.25 |
2.11. Identification of Campylobacter spp.
2.12. Statistical Analysis
3. Results
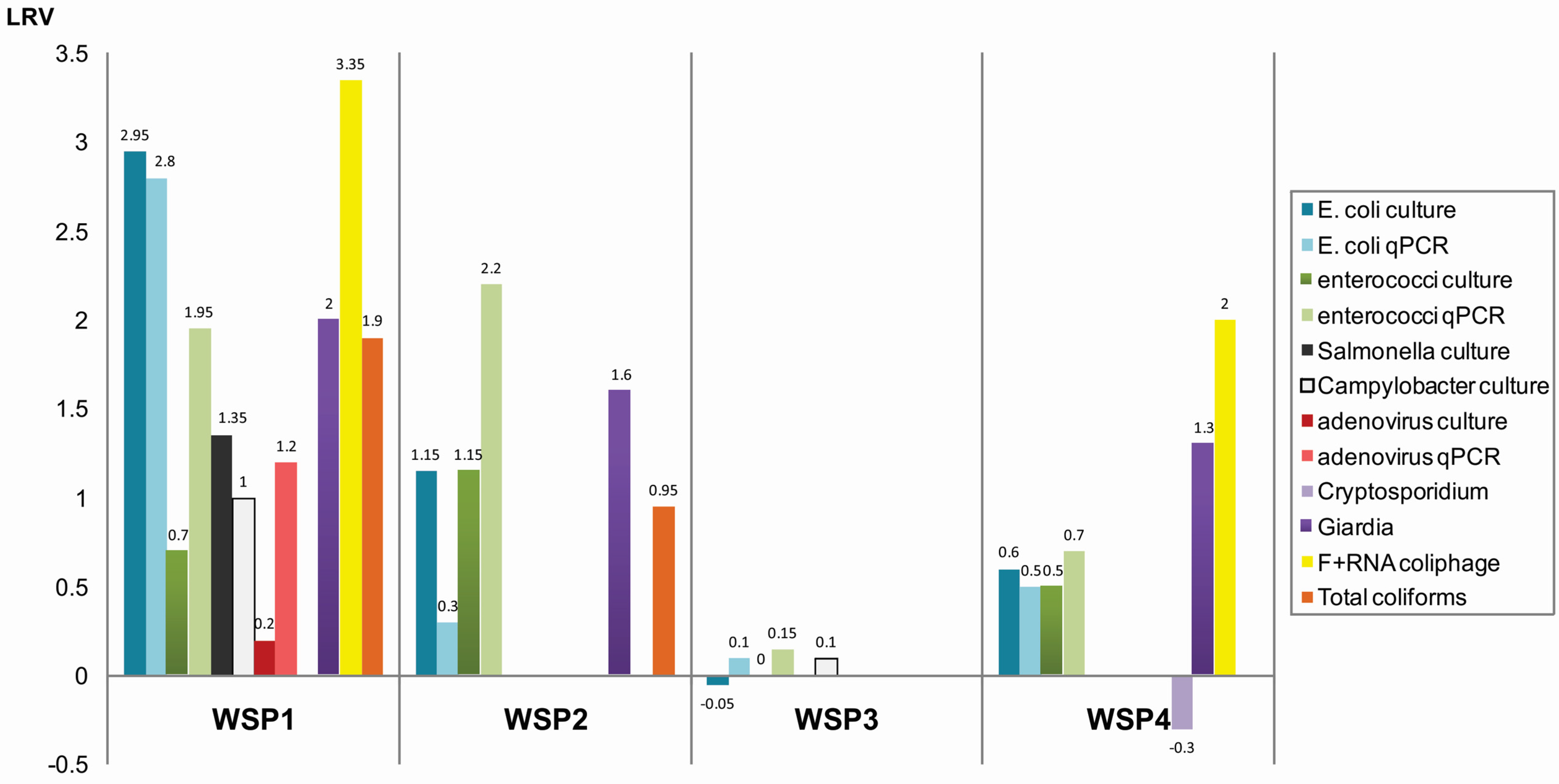
3.1. Concentrations of Microorganisms in Maturation Ponds
| Microorganism | Year | Method | WSP1, QLD (n = 8) | WSP2 *, QLD (n = 8) | WSP3, NT (n = 8) | WSP4, NT (n = 4) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Influent | Effluent | LRV | Influent | Effluent | LRV | Influent | Effluent | LRV | Influent | Effluent | LRV | |||
| E. coli | 2013 | culture | 5.6 ± 0.6 × 104 | 2.6 ± 0.7 × 101 | 3.3 | 2.8 ± 0.07 × 102 | 2.9 ± 0.4 × 102 | 0.0 | 1.9 ± 0.01 × 104 | 2.30 ± 0.01 × 104 | −0.1 | NT | NT | - |
| qPCR | 6.6 ± 3 × 104 | 2.2 ± 3.2 × 102 | 2.5 | 7.7 ± 3.3 × 103 | 3.64 ± 0.7 × 103 | 0.3 | 7.6 ± 3.3 × 105 | 4.1 ±1.12 × 105 | 0.3 | |||||
| 2014 | culture | 1.5 ± 0.2 × 105 | 3.5 ± 0.8 × 102 | 2.6 | 2 × 102 | ND | 2.3 | 1.3 ± 0.01 × 105 | 1.4 ± 0.1 × 105 | 0.0 | 1.7 ± 0.7 × 105 | 4.4 ± 0.5 × 104 | 0.6 | |
| qPCR | 1.5 ± 0.1 × 106 | 1.2 ± 6.6 × 103 | 3.1 | 3.2 ± 0.8 × 104 | ND | - | 1.7 ± 0.2 × 106 | 2.07 ± 0.1 × 106 | −0.1 | 3.1 ± 0.0 × 106 | 8.97 ± 0.06 × 105 | 0.5 | ||
| Enterococcus spp. | 2013 | culture | 1.9 ± 0.05 × 104 | 7.0 ± 4.2 × 102 | 1.4 | 7.2 ± 0.2 × 102 | 8.8 ± 1.6 × 102 | −0.1 | 4.7 × 103 | 4.3 × 103 | 0.0 | NT | NT | - |
| qPCR | 2.7 ± 1.2 × 106 | 1.9 ± 1.7 × 103 | 3.2 | 1.6 ± 0.04 × 106 | 3.4 ± 0.1 × 105 | 0.7 | 3.2 ± 1.05 × 106 | 2.16 ± 1.32 × 106 | 0.2 | - | ||||
| 2014 | culture | 4.4± 0.07 × 104 | 4.3 ± 0.6 × 104 | 0.0 | 3.0 × 102 | ND | 2.4 | 1.1 ± 0.6 × 103 | 1.1 ± 0.3 × 103 | ND | 2.8 ± 0.6 × 104 | 9.1 ± 5.3 × 103 | 0.5 | |
| qPCR | 1.5 × 107 | 3 × 106 | 0.7 | 4.4 ± 0.6 × 106 | 9.1 ± 2.2 × 102 | 3.7 | 1.9 ± 1.01 × 104 | 1.5 ± 0.21 × 104 | 0.1 | 2.5 ± 0.01 × 105 | 4.9 ± 0.7 × 104 | 0.7 | ||
| Salmonella spp. | 2013 | culture | 3.0 ± 0.4 ×103 | 9.5 ± 7.8 × 101 | 1.5 | 3.2 ± 0.2 × 102 | 4.1 ± 0.8 × 102 | −0.1 | NT | NT | - | NT | NT | - |
| qPCR | ND | ND | - | ND | ND | - | ND | ND | - | |||||
| 2014 | culture | 5.5 ± 1.5 × 104 | 3.4 ± 1.1 × 103 | 1.2 | ND | ND | 0 | NT | NT | - | NT | NT | - | |
| qPCR | ND | ND | - | ND | ND | - | - | |||||||
| Campylobacter spp. | 2013 | qPCR | ND | ND | - | ND | ND | - | ND | ND | - | NT | NT | - |
| 2014 | culture | 7.0 × 101 | 6.6 ± 5.7 | 1.0 | ND | ND | - | 9.6 ± 0.1 × 103 | 8.0 ± 1.3 × 103 | 0.1 | NT | NT | - | |
| qPCR | ND | ND | - | ND | ND | - | ND | ND | - | ND | ND | |||
| adenovirus | 2014 | qPCR | 4.8 ± 1.5 × 103 | 3.03 ± 1.3 × 103 | 0.2 | ND | ND | - | 2.0 × 105 | 2.4 × 105 | −0.1 | NT | 12.04 MPNIU | - |
| 2015 | culture | 23 MPNIU | 6.6 MPNIU | 1.2 | ||||||||||
| Cryptosporidium spp. | 2014 | microscopy | 0.5 ± 0.7 (31%) | ND (22 %) | - | ND (16%) | ND (30%) | - | NT | NT | - | 2 ± 1.7 (3%) | 2.5 ± 1.4 (20%) | −0.3 |
| 2015 | microscopy | 0.5 ± 0.7 (25%) | ND (24%) | - | NT | NT | ||||||||
| Giardia spp. | 2014 | microscopy | 119 ± 130 (21%) | 1 ± 1.4 (11%) | 2.2 | 7.5 (18%) | ND (30%) | 1.6 | NT | NT | - | 329.9 ± 7.6 (3%) | 154.7 ± 30.7 (30%) | 1.3 |
| 2015 | microscopy | 76.5 ± 64.3 (26%) | ND (14%) | 1.8 | ||||||||||
| F+ RNA coliphage | 2013 | culture | 1.0 × 107 ± 2.0 | 0 | n/a | ND | ND | - | NT | NT | - | NT | NT | - |
| 2014 | culture | <1.0 × 105 | 2.0 × 101 | 3.7 | ND | ND | - | NT | NT | - | 3.4 ± 4.0 × 103 | 3.6 ± 4.9 × 101 | 2.0 | |
| 2015 | culture | 9 × 103 | 13 | 3.0 | ||||||||||
| Total coliforms | 2013 | culture | 3.4 ± 0.2 × 104 | 4.1 × 102 | 1.9 | 8.8 ± 1.3 × 102 | 5.9 ± 1.2 × 102 | 0.2 | NT | NT | - | NT | NT | - |
| 2014 | culture | 2.0 ± 0.1 × 105 | 2.4 ± 0.2 × 103 | 1.9 | 7.6 ± 0.2 × 103 | 1.5 ± 1.0 × 102 | 1.7 | NT | NT | - | NT | NT | - | |
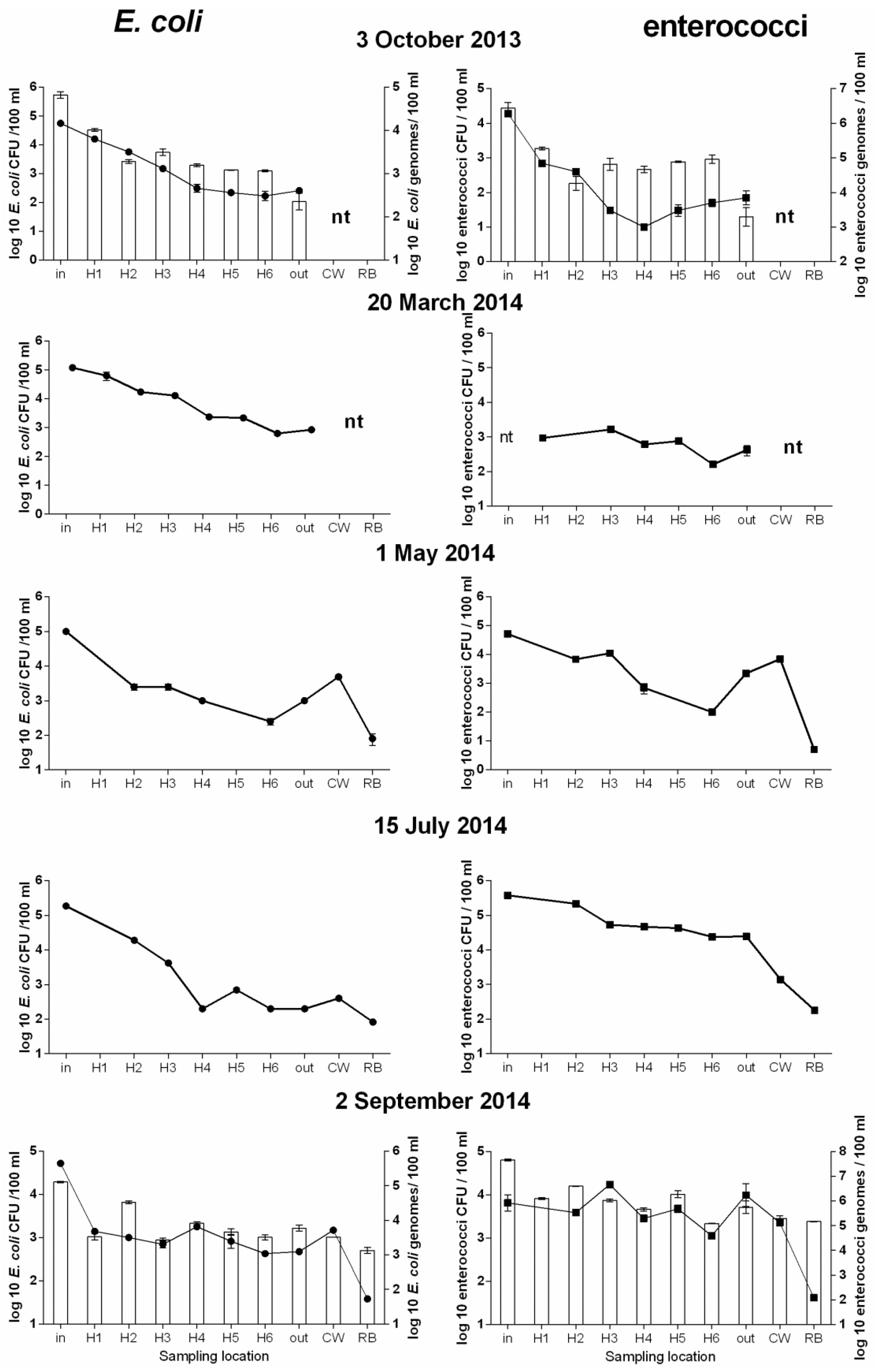
3.2. Further Analysis of Microbial Concentration of WSP1
4. Discussion
4.1. Faecal Indicators and Pathogen Concentrations
4.2. Comparison of qPCR Data and Culture Methods for Fecal Indicators
4.3. Salmonella spp. and Campylobacter spp. Enumeration
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mara, D. Waste stabilization ponds: Past, present and future. Desalin. Water Treat. 2009, 4, 85–88. [Google Scholar] [CrossRef]
- Mara, D. Constructed wetlands are not a viable alternative or addition to waste stabilization ponds. In Proceeding of the 7th IWA Specialist Conference On Waste Stabilization Ponds, Bangkok, Thailand, 25–27 September 2006.
- Greenway, M. The role of constructed wetlands in secondary effluent treatment and water reuse in subtropical and arid australia. Ecol. Eng. 2005, 25, 501–509. [Google Scholar] [CrossRef]
- Higgins, J.; Warnken, J.; Teasdale, P.R. Quality criteria in australian reclaimed water guidelines and effluent discharge licences: How consistent are we? Australas. J. Environ. Manag. 2004, 11, 227–236. [Google Scholar] [CrossRef]
- Ahmed, W.; Sawant, S.; Huygens, F.; Goonetilleke, A.; Gardner, T. Prevalence and occurrence of zoonotic bacterial pathogens in surface waters determined by quantitative PCR. Water Res. 2009, 43, 4918–4928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellein, K.N.; Battie, C.; Tauchman, E.; Lund, D.; Oyarzabal, O.A.; Lepo, J.E. Culture-based indicators of fecal contamination and molecular microbial indicators rarely correlate with Campylobacter spp. in recreational waters. J. Water Health 2011, 9, 695–707. [Google Scholar] [CrossRef] [PubMed]
- NRMMC-EPHC-AHMC. Australian Guidelines for Water Recycling: Managing Health and Environmental Risks (Phase 2): Augmentation of Drinking Water Supplies; NRMMC-EPHC-AHMC: Sydney, Australia, 2008; Volume Draft for Public Comment. [Google Scholar]
- APHA. Standard Methods for the Examination of Water and Wastewater, 18th ed.; American Public Health Assosiation: Washington, DC, USA, 1992. [Google Scholar]
- Fravalo, P.; Hascoet, Y.; Fellic, M.L.; Queguiner, S.; Petton, J.; Salvat, G. Convenient method for rapid and quantitative assessment of Salmonella enterica contamination: The Mini-MSRV MPN technique. J. Rapid Methods Autom. Microbiol. 2003, 11, 81–88. [Google Scholar] [CrossRef]
- Bosshard, F.; Armand, F.; Hamelin, R.; Kohn, T. Mechanisms of human adenovirus inactivation by sunlight and UVC light as examined by quantitative pcr and quantitative proteomics. Appl. Environ. Microbiol. 2013, 79, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- IDT PrimerDesign. Available online: http://www.idtdna.com/primerquest/home/index (accessed on 31 December 2015).
- GenBank BlastN. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 31 December 2015).
- Usachev, E.V.; Agranovski, I.E. Internally controlled pcr system for detection of airborne microorganisms. J. Environ. Monit. 2012, 14, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Frahm, E.; Obst, U. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 2003, 52, 123–131. [Google Scholar] [CrossRef]
- USEPA. Method A: Enterococci in Water by TaqMan Quantitative Polymerase Chain Reaction (qPCR) Assay; Office of Water: Washington, DC, USA, 2012. [Google Scholar]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Åkerblom, P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003, 70, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Inglis, G.D.; Kalischuk, L.D. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time quantitative PCR. Appl. Environ. Microbiol. 2004, 70, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.; Stratton, H.M.; Schreoder, S.; Toze, S. Pathogen Detection Technologies for Wastewater and Reservoirs; Urban Water Security Research Alliance Technical Report No. 32; UWSRA: Brisbane, Australia, 2010. [Google Scholar]
- Yang, C.; Jiang, Y.; Huang, K.; Zhu, C.; Yin, Y. Application of real-time PCR for quantitative detection of Campylobacter jejuni in poultry, milk and environmental water. FEMS Immunol. Med. Microbiol. 2003, 38, 265–271. [Google Scholar] [CrossRef]
- Leo, A.; Tow, W.; Hill, J.; English, J.; Ford, R. Novel Salmonella spp. diagnostic markers based on the gyrB gene and its application on food and environmentally-delivered water samples. J. Adv. Biotechnol. Bioeng. 2013, 1, 30–39. [Google Scholar]
- Jakočiūnė, D.; Pasquali, F.; da Silva, C.S.; Löfström, C.; Hoorfar, J.; Klein, G.; Manfreda, G.; Olsen, J.E. Enumeration of salmonellae in table eggs, pasteurized egg products, and egg-containing dishes by using quantitative real-time PCR. Appl. Environ. Microbiol. 2014, 80, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Denis, M.; Soumet, C.; Rivoal, K.; Ermel, G.; Blivet, D.; Salvat, G.; Colin, P. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 1999, 29, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Ouali, A.; Jupsin, H.; Vasel, J.L.; Marouani, L.; Ghrabi, A. Removal improvement of bacteria (Escherichia coli and Enterococci) in maturation ponds using baffles. Water Sci. Technol. 2012, 65, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Shilton, A. Pond Treatment Technology; IWA Publishing: London, UK, 2005. [Google Scholar]
- Brooks, A.; Antenucci, J.; Hipsey, M.; Burch, M.D.; Ashbolt, N.J.; Ferguson, C. Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 2004, 30, 741–759. [Google Scholar] [CrossRef] [PubMed]
- Kohn, T.; Nelson, K.L. Sunlight-mediated inactivation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters. Environ. Sci. Technol. 2006, 41, 192–197. [Google Scholar] [CrossRef]
- McFeters, G.A.; Bissonnette, G.K.; Jezeski, J.J.; Thomson, C.A.; Stuart, D.G. Comparative survival of indicator bacteria and enteric pathogens in well water. Appl. Microbiol. 1974, 27, 823–829. [Google Scholar] [PubMed]
- Edberg, S.C.; Leclerc, H.; Robertson, J. Natural protection of spring and well drinking water against surface microbial contamination. II. Indicators and monitoring parameters for parasites. Crit. Rev. Microbiol. 1997, 23, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Davies-Colley, R.J.; Donnison, A.M.; Speed, D.J.; Ross, C.M.; Nagels, J.W. Inactivation of faecal indicator micro-organisms in waste stabilisation ponds: Interactions of environmental factors with sunlight. Water Res. 1999, 33, 1220–1230. [Google Scholar] [CrossRef]
- Kadir, K.; Nelson, K.L. Sunlight mediated inactivation mechanisms of Enterococcus faecalis and Escherichia coli in clear water vs. waste stabilization pond water. Water Res. 2014, 50, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Jasper, J.T.; Boehm, A.B.; Nelson, K.L. Sunlight inactivation of fecal indicator bacteria in open-water unit process treatment wetlands: Modeling endogenous and exogenous inactivation rates. Water Res. 2015, 83, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, J.P.S.; Ahmed, W.; Toze, S. Sensitive detection of human adenovirus from small volume of primary wastewater samples by quantitative PCR. J. Virol. Methods 2013, 187, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.J.; Smith, P.G.; Grimason, A.T.; Smith, H.V. Removal and destruction of intestinal parasitic protozoans by sewage treatment processes. Int. J. Environ. Health Res. 1999, 9, 85–96. [Google Scholar] [CrossRef]
- Reinoso, R.; Blanco, S.; Torres-Villamizar, L.A.; Becares, E. Mechanisms for parasites removal in a waste stabilisation pond. Microb. Ecol. 2011, 61, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Wohlsen, T.; Bates, J.; Gray, B.; Aldridge, P.; Stewart, S.; Williams, M.; Katouli, M. The occurrence of Cryptosporidium and Giardia in the Lake Baroon catchment, Queensland, Australia. J. Water Supply Res. Technol. 2006, 55, 357–366. [Google Scholar]
- Wéry, N.; Lhoutellier, C.; Ducray, F.; Delgenès, J.-P.; Godon, J.-J. Behaviour of pathogenic and indicator bacteria during urban wastewater treatment and sludge composting, as revealed by quantitative PCR. Water Res. 2008, 42, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. Problems in Detecting Dormant (VBNC) Cells, and the Role of DNA Elements in This Response; Landes Bioscience: Georgetown, DC, USA, 2000; pp. 1–15. [Google Scholar]
- Van Frankenhuyzen, J.K.; Trevors, J.T.; Lee, H.; Flemming, C.A.; Habash, M.B. Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration. J. Microbiol. Method. 2011, 87, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Cappelier, J.M.; Minet, J.; Magras, C.; Colwell, R.R.; Federighi, M. Recovery in embryonated eggs of viable but nonculturable Campylobacter jejuni cells and maintenance of ability to adhere to hela cells after resuscitation. Appl. Environ. Microbiol. 1999, 65, 5154–5157. [Google Scholar] [PubMed]
- Lebuhn, M.; Effenberger, M.; Garces, G.; Gronauer, A.; Wilderer, P.A. Hygienization by anaerobic digestion: Comparison between evaluation by cultivation and quantitative real-time PCR. Water Sci. Technol. 2005, 52, 93–99. [Google Scholar] [PubMed]
- England, L.S.; Pollok, J.; Vincent, M.; Kreutzweiser, D.; Fick, W.; Trevors, J.T.; Holmes, S.B. Persistence of extracellular baculoviral DNA in aquatic microcosms: Extraction, purification, and amplification by the polymerase chain reaction (PCR). Mol. Cell. Probes 2005, 19, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Botes, M.; De Kwaadsteniet, M.; Cloete, T.E. Application of quantitative PCR for the detection of microorganisms in water. Anal. Bioanal. Chem. 2013, 405, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Hussong, D.; Enkiri, N.K.; Burge, W.D. Modified agar medium for detecting environmental salmonellae by the most-probable-number method. Appl. Environ. Microbiol. 1984, 48, 1026–1030. [Google Scholar] [PubMed]
- Kumar, R.; Surendran, P.K.; Thampuran, N. Rapid quantification of salmonella in seafood by real-time PCR assay. J. Microbiol. Biotechnol. 2010, 20, 569–573. [Google Scholar] [PubMed]
- Shannon, K.E.; Lee, D.Y.; Trevors, J.T.; Beaudette, L.A. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 2007, 382, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Oragui, J.I.; Curtis, T.P.; Silva, S.A.; Mara, D.D. The removal of excreted bacteria and viruses in deep waste stabilization ponds in northeast Brazil. Water Sci. Technol. 1987, 19, 569–573. [Google Scholar]
- Chon, J.-W.; Hyeon, J.-Y.; Yim, J.-H.; Kim, J.-H.; Song, K.-Y.; Seo, K.-H. Improvement of modified charcoal-cefoperazone-deoxycholate agar by supplementation with a high concentration of polymyxin B for detection of Campylobacter jejuni and C. coli in chicken carcass rinses. Appl. Environ. Microbiol. 2012, 78, 1624–1626. [Google Scholar] [CrossRef] [PubMed]
- Engberg, J.; On, S.L.; Harrington, C.S.; Gerner-Smidt, P. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for Campylobacters. J. Clin. Microbiol. 2000, 38, 286–291. [Google Scholar] [PubMed]
- Clark, S.T.; Gilbride, K.A.; Mehrvar, M.; Laursen, A.E.; Bostan, V.; Pushchak, R.; McCarthy, L.H. Evaluation of low-copy genetic targets for waterborne bacterial pathogen detection via qPCR. Water Res. 2011, 45, 3378–3388. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.H.; Gannon, V.; Kent, R.; Koning, W.; Lapen, D.R.; Miller, J.; Neumann, N.; Phillips, R.; Robertson, W.; Topp, E.; et al. Development of a rapid quantitative PCR assay for direct detection and quantification of culturable and non-culturable Escherichia coli from agriculture watersheds. J. Microbiol. Method. 2007, 69, 480–488. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheludchenko, M.; Padovan, A.; Katouli, M.; Stratton, H. Removal of Fecal Indicators, Pathogenic Bacteria, Adenovirus, Cryptosporidium and Giardia (oo)cysts in Waste Stabilization Ponds in Northern and Eastern Australia. Int. J. Environ. Res. Public Health 2016, 13, 96. https://doi.org/10.3390/ijerph13010096
Sheludchenko M, Padovan A, Katouli M, Stratton H. Removal of Fecal Indicators, Pathogenic Bacteria, Adenovirus, Cryptosporidium and Giardia (oo)cysts in Waste Stabilization Ponds in Northern and Eastern Australia. International Journal of Environmental Research and Public Health. 2016; 13(1):96. https://doi.org/10.3390/ijerph13010096
Chicago/Turabian StyleSheludchenko, Maxim, Anna Padovan, Mohammad Katouli, and Helen Stratton. 2016. "Removal of Fecal Indicators, Pathogenic Bacteria, Adenovirus, Cryptosporidium and Giardia (oo)cysts in Waste Stabilization Ponds in Northern and Eastern Australia" International Journal of Environmental Research and Public Health 13, no. 1: 96. https://doi.org/10.3390/ijerph13010096







