Leptospira spp. in Rodents and Shrews in Germany
Abstract
:1. Introduction
2. Experimental Section
3. Results
3.1. Description of Examined Animals
| Federal State | No. of Regions | Total No. of Animals | Median (Range) Animals/Region |
|---|---|---|---|
| Brandenburg | 14 | 1065 | 77 (1–241) |
| Baden-Württemberg | 9 | 202 | 15 (1–76) |
| Bavaria | 6 | 107 | 13 (5–45) |
| Mecklenburg-West Pomerania | 12 | 758 | 20 (5–329) |
| Lower Saxony | 6 | 377 | 15 (1–257) |
| North Rhine-Westphalia | 6 | 226 | 8 (1–83) |
| Hesse | 1 | 32 | - |
| Rhineland-Palatinate | 1 | 6 | - |
| Saxony | 1 | 13 | - |
| Saxony-Anhalt | 2 | 150 | - |
| Schleswig-Holstein | 2 | 34 | - |
| Total | 60 | 2970 * | - |
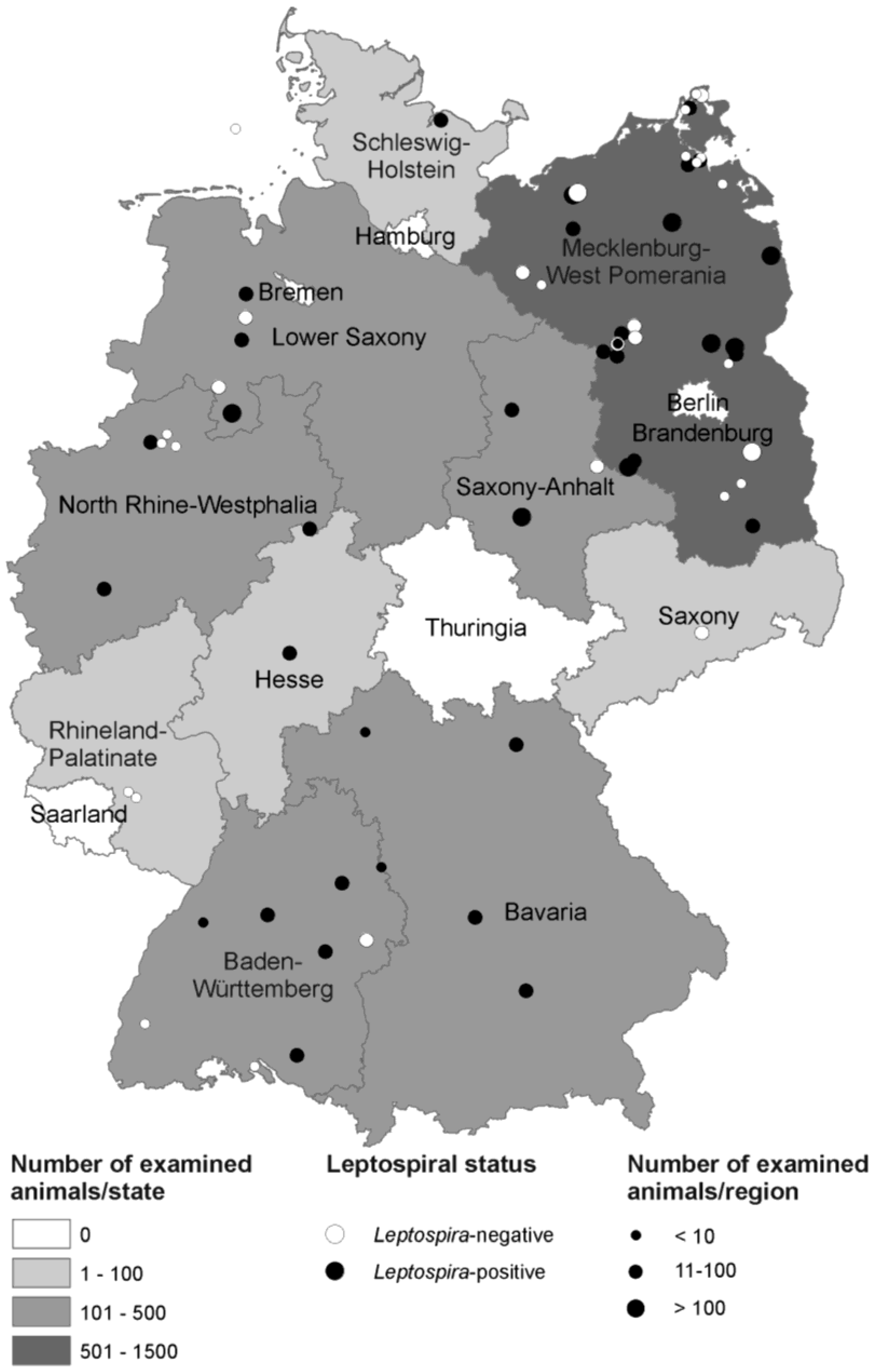
3.2. Identification of Leptospira spp. in Trapped Rodents and Shrews
| Rodents and Shrews | Leptospira-positive Animals ** | Number of Identified Leptospira spp. *** | |||||
|---|---|---|---|---|---|---|---|
| Genus | Species | No. Trapped | Total Number | Percentage | L. kirschneri | L. interrogans | L. borgpetersenii |
| Apodemus | agrarius | 190 | 23 | 12 | 19 | 0 | 0 |
| flavicollis | 792 | 71 | 9 | 17 | 36 | 1 | |
| sylvaticus | 154 | 27 | 18 | 6 | 13 | 0 | |
| spp. total | 1136 | 121 | 11 | 42 | 49 | 1 | |
| Micromys | minutus | 6 | 0 | 0 | 0 | 0 | 0 |
| Mus | musculus | 18 | 1 | 6 | 1 | 0 | 0 |
| Arvicola | amphibius | 8 | 2 | 25 | 2 | 0 | 0 |
| Clethrionomys | glareolus | 1016 | 66 | 6 | 38 | 18 | 0 |
| Microtus | agrestis | 517 | 64 | 12 | 38 | 1 | 6 |
| arvalis | 174 | 24 | 14 | 19 | 0 | 0 | |
| oeconomus | 2 | 1 | 50 | 1 | 0 | 0 | |
| subterraneus | 4 | 0 | 0 | 0 | 0 | 0 | |
| spp. total | 697 | 89 | 13 | 58 | 1 | 6 | |
| Crocidura | leucodon | 3 | 1 | 33 | 0 | 0 | 0 |
| russula | 24 | 5 | 21 | 5 | 0 | 0 | |
| suaveolens | 3 | 0 | 0 | 0 | 0 | 0 | |
| spp. total | 30 | 6 | 20 | 5 | 0 | 0 | |
| Sorex | araneus | 36 | 2 | 6 | 0 | 0 | 0 |
| coronatus | 2 | 1 | 50 | 1 | 0 | 0 | |
| minutus | 12 | 0 | 0 | 0 | 0 | 0 | |
| spp. total | 50 | 3 | 6 | 1 | 0 | 0 | |
| Total | 2961 * | 288 | 10 | 147 | 68 | 7 | |
3.3. Detailed Description of Leptospira Detection in Selected Murine and Vole Species
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; Vinetz, J.M. Peru—United States Leptospirosis Consortium. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef]
- Faine, S.; Adler, B.; Bolin, C.; Perolat, P. Leptospira and Leptospirosis; MediSci: Melbourne, Australia, 1999; pp. 83–86. [Google Scholar]
- Picardeau, M. Diagnosis and epidemiology of leptospirosis. Med. Mal. Infec. 2013, 43, 1–9. [Google Scholar] [CrossRef]
- Robert Koch Institute. Available online: https://www3.rki.de/survstat/ (accessed on 15 July 2014).
- Jansen, A.; Schöneberg, I.; Frank, C.; Alpers, K.; Schneider, T.; Stark, K. Leptospirosis in Germany 1962–2003. Emerg. Infect. Dis. 2005, 11, 1048–1054. [Google Scholar] [CrossRef]
- Desai, S.; van Treeck, U.; Lierz, M.; Espelage, W.; Zota, L.; Sarbu, A.; Czerwinski, M.; Sadkowska-Todys, M.; Avdicová, M.; Reetz, J.; Luge, E.; Guerra, B.; Nöckler, K.; Jansen, A. Resurgence of field fever in a temperate country: An epidemic of leptospirosis among seasonal strawberry harvesters in Germany in 2007. Clin. Infect. Dis. 2009, 48, 691–697. [Google Scholar] [CrossRef]
- van Crevel, R.; Speelman, P.; Gravekamp, C.; Terpstra, W.J. Leptospirosis in travellers. Clin. Infect. Dis. 1994, 19, 132–134. [Google Scholar] [CrossRef]
- Sejvar, J.; Bancroft, E.; Winthrop, K.; Bettinger, J.; Bajani, M.; Bragg, S.; Shutt, K.; Kaiser, R.; Marano, N.; Popovic, T.; Tappero, J.; Ashford, D.; Mascola, L.; Vugia, D.; Perkins, B.; Rosenstein, N. Eco-challenge investigation team. Leptospirosis in “eco-challenge” athletes, Malaysian Borneo, 2000. Emerg. Infect. Dis. 2003, 9, 702–707. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Hartskeerl, R.A.; Terpstra, W.J. Leptospirosis in wild animals. Vet. Quart. 1996, 18, S149–S150. [Google Scholar] [CrossRef]
- Monahan, A.M.; Callanan, J.J.; Nally, J.E. Host-pathogen interactions in the kidney during chronic leptospirosis. Vet. Pathol. 2009, 46, 792–799. [Google Scholar] [CrossRef]
- Mitchell-Jones, J.; Amori, G.; Bogdanowicz, W.; Krystufek, B.; Reijnders, P.J.H.; Spitzenberger, F.; Stubbe, M.; Thissen, J.B.M.; Vohralik, V.; Zima, J. The Atlas of European Mammals; T & AD Poyser, Academic Press: London, UK, 1999; pp. 32–311. [Google Scholar]
- Essbauer, S.S.; Schmidt-Chanasit, J.; Madeja, E.L.; Wegener, W.; Friedrich, R.; Petraityte, R.; Sasnauskas, K.; Jacob, J.; Koch, J.; Dobler, G.; Conraths, F.J.; Pfeffer, M.; Pitra, C.; Ulrich, R.G. Nephropathia epidemica outbreak in a metropolitan area, Germany. Emerg. Infect. Dis. 2007, 13, 1271–1273. [Google Scholar]
- Ehlers, B.; Kuchler, J.; Yasmum, N.; Dural, G.; Schmidt-Chanasit, J.; Jäkel, T.; Matuschka, F.-R.; Richter, D.; Essbauer, S.; Hughes, D.J.; Summers, C.; Bennett, M.; Stewart, J.P.; Ulrich, R.G. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J. Virol. 2007, 81, 8091–8100. [Google Scholar] [CrossRef]
- Guenther, S.; Grobbel, M.; Heidemanns, K.; Schlegel, M.; Ulrich, R.G.; Ewers, C.; Wieler, L.H. First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Total Envir. 2010, 408, 3519–3522. [Google Scholar]
- Schmidt-Chanasit, J.; Essbauer, S.; Petraityte, R.; Yoshimatsu, K.; Tackmann, K.; Conraths, F.J.; Sasnauskas, K.; Arikawa, J.; Thomas, A.; Pfeffer, M.; Scharninghausen, J.J.; Splettstoesser, W.; Wenk, M.; Heckel, G.; Ulrich, R.G. Extensive host sharing of central European Tula virus. J. Virol. 2010, 84, 459–474. [Google Scholar] [CrossRef]
- Ettinger, J.; Hofmann, J.; Enders, M.; Tewes, F.; Oehme, R.M.; Rosenfeld, U.M.; Ali, H.S.; Schlegel, M.; Essbauer, S.; Osterberg, A.; Jacob, J.; Reil, D.; Klempa, B.; Ulrich, R.G.; Krüger, D.H. Multiple synchronous Puumala virus outbreaks, Germany, 2010. Emerg. Infect. Dis. 2012, 18, 1461–1464. [Google Scholar] [CrossRef]
- Schlegel, M.; Radosa, L.; Rosenfeld, U.M.; Schmidt, S.; Triebenbacher, C.; Löhr, P.-W.; Fuchs, D.; Heroldová, M.; Jánová, E.; Stanko, M.; Mošanský, L.; Fričová, J.; Pejčoch, M.; Suchomel, J.; Purchart, L.; Groschup, M.H.; Krüger, D.H.; Klempa, B.; Ulrich, R.G. Broad geographical distribution and high genetic diversity of shrew-borne Seewis hantavirus in Central Europe. Virus Genes 2012, 45, 48–55. [Google Scholar] [CrossRef]
- Schlegel, M.; Klempa, B.; Auste, B.; Bemmann, M.; Schmidt-Chanasit, J.; Büchner, T.; Groschup, M.H.; Meier, M.; Buschmann, A.; Zoller, H.; Krüger, D.H.; Ulrich, R.G. Multiple dobrava-belgrade virus spillover infections, Germany. Emerg. Infect. Dis. 2009, 15, 2017–2020. [Google Scholar] [CrossRef]
- Hofmann, J.; Meisel, H.; Klempa, B.; Vesenbeckh, S.M.; Beck, R.; Michel, D.; Schmidt-Chanasit, J.; Ulrich, R.G.; Grund, S.; Enders, G.; Krüger, D.H. Hantavirus outbreak, Germany, 2007. Emerg. Infect. Dis. 2008, 14, 850–852. [Google Scholar] [CrossRef]
- De Melo, V.W.; Ali, H.S.; Freise, J.; Essbauer, S.; Mertens, M.; Wanka, K.; Drewes, S.; Ulrich, R.G.; Heckel, G. Spatiotemporal dynamics of Puumala hantavirus associated with its rodent host, myodes glareolus. Mol. Ecol. 2014. submitted. [Google Scholar]
- Ulrich, R.G.; Schmidt-Chanasit, J.; Schlegel, M.; Jacob, J.; Pelz, H.J.; Mertens, M.; Wenk, M.; Büchner, T.; Masur, D.; Sevke, K.; Groschup, M.H.; Gerstengarbe, F.W.; Pfeffer, M.; Oehme, R.; Wegener, W.; Bemmann, M.; Ohlmeyer, L.; Wolf, R.; Zoller, H.; Koch, J.; Brockmann, S.; Heckel, G.; Essbauer, S.S. Network “rodent-borne pathogens” in Germany: Longitudinal studies on the geographical distribution and prevalence of hantavirus infections. Parasitol. Res. 2008, 103, S121–S129. [Google Scholar] [CrossRef]
- Schlegel, M.; Ali, H.S.; Stieger, N.; Groschup, M.H.; Wolf, R.; Ulrich, R.G. Molecular identification of small mammal species using novel cytochrome b gene-derived degenerated primers. Biochem. Genet. 2012, 50, 440–447. [Google Scholar] [CrossRef]
- Gravekamp, C.; van de Kemp, H.; Franzen, M.; Carrington, D.; Schoone, G.J.; van Eys, G.J.; Everard, C.O.; Hartskeerl, R.A.; Terpstra, W.J. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J. Gen. Microbiol. 1993, 139, 1691–1700. [Google Scholar] [CrossRef]
- Mayer-Scholl, A.; Draeger, A.; Luge, E.; Ulrich, R.; Nöckler, K. Comparison of two PCR systems for the rapid detection of Leptospira. spp. from kidney tissue. Curr. Microbiol. 2011, 62, 1104–1106. [Google Scholar] [CrossRef]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira. spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Slack, A.T.; Symonds, M.L.; Dohnt, M.F.; Smythe, L.D. Identification of pathogenic Leptospira. species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006, 6. [Google Scholar] [CrossRef]
- Victoria, B.; Ahmed, A.; Zuerner, R.L.; Ahmed, N.; Bulach, D.M.; Quinteiro, J.; Hartskeerl, R.A. Conservation of the S10-spc α locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One 2008. [Google Scholar] [CrossRef]
- Runge, M.; von Keyserlingk, M.; Braune, S.; Becker, D.; Plenge-Bönig, A.; Freise, J.F.; Pelz, H.J.; Esther, A. Distribution of rodenticide resistance and zoonotic pathogens in Norway rats in Lower Saxony and Hamburg, Germany. Pest. Manag. Sci. 2012, 69, 403–408. [Google Scholar]
- Aviat, F.; Blanchard, B.; Michel, V.; Blanchet, B.; Branger, C.; Hars, J.; Mansotte, F.; Brasme, L.; de Champs, C.; Bolut, P.; Mondot, P.; Faliu, J.; Rochereau, S.; Kodjo, A.; Andre-Fontaine, G. Leptospira. exposure in the human environment in France: A survey in feral rodents and in fresh water. Comp. Immunol. Microbiol. Infect. 2009, 32, 463–476. [Google Scholar] [CrossRef]
- Theuerkauf, J.; Perez, J.; Taugamoa, A.; Niutoua, I.; Labrousse, D.; Gula, R.; Bogdanowicz, W.; Jourdan, H.; Goarant, C. Leptospirosis risk increases with changes in species composition of rat populations. Naturwissenschaften 2013, 100, 385–388. [Google Scholar] [CrossRef]
- Kocianová, E.; Kozuch, O.; Bakoss, P.; Rehácek, J.; Kovácová, E. The prevalence of small terrestrial mammals infected with tick-borne encephalitis virus and leptospirae in the foothills of the southern Bavarian forest, Germany. Appl. Parasitol. 1993, 34, 283–290. [Google Scholar]
- Woll, D.; Karnath, C.; Pfeffer, M.; Allgöwer, R. Genetic characterization of Leptospira spp. from beavers found dead in south-west Germany. Vet. Microbiol. 2012, 158, 232–234. [Google Scholar] [CrossRef]
- Adler, H.; Vonstein, S.; Deplazes, P.; Stieger, C.; Frei, R. Prevalence of Leptospira. spp. in various species of small mammals caught in an inner-city area in Switzerland. Epidemiol. Infect. 2002, 128, 107–109. [Google Scholar]
- Turk, N.; Milas, Z.; Margaletic, J.; Staresina, V.; Slavica, A.; Riquelme-Sertour, N.; Bellenger, E.; Baranton, G.; Postic, D. Molecular characterization of Leptospira. spp. strains isolated from small rodents in Croatia. Epidemiol. Infect. 2003, 130, 159–166. [Google Scholar] [CrossRef]
- Schmidt, S.; Essbauer, S.S.; Mayer-Scholl, A.; Poppert, S.; Schmidt-Chanasit, J.; Klempa, B.; Henning, K.; Schares, G.; Groschup, M.H.; Spitzenberger, F.; Heckel, G.; Richter, D.; Ulrich, R.G. Multiple infections of rodents with zoonotic pathogens in Austria. Vector-Borne Zoonotic Dis. 2014, 14, 467–475. [Google Scholar]
- Baker, P.J.; Ansell, R.J.; Dodds, P.A.A.; Webber, C.E.; Harris, S. Factors affecting the distribution of small mammals in an urban area. Mammal Rev. 2003, 33, 95–100. [Google Scholar] [CrossRef]
- Dickman, C.R.; Doncaster, C.P. The ecology of small mammals in urban habitats. I. Populations in a patchy environment. J. Anim. Ecol. 1987, 56, 629–640. [Google Scholar] [CrossRef]
- Tattersall, F.H. House mice and wood mice in and around an agricultural building. J. Zool. Lond. 1999, 249, 469–472. [Google Scholar] [CrossRef]
- Dupouey, J.; Faucher, B.; Edouard, S.; Richet, H.; Kodjo, A.; Drancourt, M.; Davoust, B. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. 2014, 37, 77–83. [Google Scholar] [CrossRef]
- Mochmann, H. The role of the field hamster (Cricetus. cricetus) as of a source of infection of the swamp-field fever. Z. Hyg. Infektionskr. 1957, 143, 327–333. [Google Scholar] [CrossRef]
- Sauer, J.R.; Slade, N.A. Size-based demography of vertebrates. Annu. Rev. Ecol. Syst. 1987, 18, 71–90. [Google Scholar]
- Paiva-Cardoso, M.N.; Collares-Pereira, M.; Gilmore, C.; Dunbar, K.; Frizzel, C.; Santos-Reis, M.; Vale-Goncalves, H.; Rodrigues, J.; Mackie, D.; Ellis, W.A. Sexual Maturity State as Risk Factor to Leptospira. Infection in Small Mammals in Tras-os-Montes e Alto Douro Region, Northern Portugal. In Proceedings of the European Meeting of Leptospirosis, Eurolepto 2012, Dubrovnik, Croatia, 31 May–2 June 2012.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mayer-Scholl, A.; Hammerl, J.A.; Schmidt, S.; Ulrich, R.G.; Pfeffer, M.; Woll, D.; Scholz, H.C.; Thomas, A.; Nöckler, K. Leptospira spp. in Rodents and Shrews in Germany. Int. J. Environ. Res. Public Health 2014, 11, 7562-7574. https://doi.org/10.3390/ijerph110807562
Mayer-Scholl A, Hammerl JA, Schmidt S, Ulrich RG, Pfeffer M, Woll D, Scholz HC, Thomas A, Nöckler K. Leptospira spp. in Rodents and Shrews in Germany. International Journal of Environmental Research and Public Health. 2014; 11(8):7562-7574. https://doi.org/10.3390/ijerph110807562
Chicago/Turabian StyleMayer-Scholl, Anne, Jens Andre Hammerl, Sabrina Schmidt, Rainer G. Ulrich, Martin Pfeffer, Dietlinde Woll, Holger C. Scholz, Astrid Thomas, and Karsten Nöckler. 2014. "Leptospira spp. in Rodents and Shrews in Germany" International Journal of Environmental Research and Public Health 11, no. 8: 7562-7574. https://doi.org/10.3390/ijerph110807562






