Oleic Acid Produced by a Marine Vibrio spp. Acts as an Anti-Vibrio parahaemolyticus Agent
Abstract
:1. Introduction
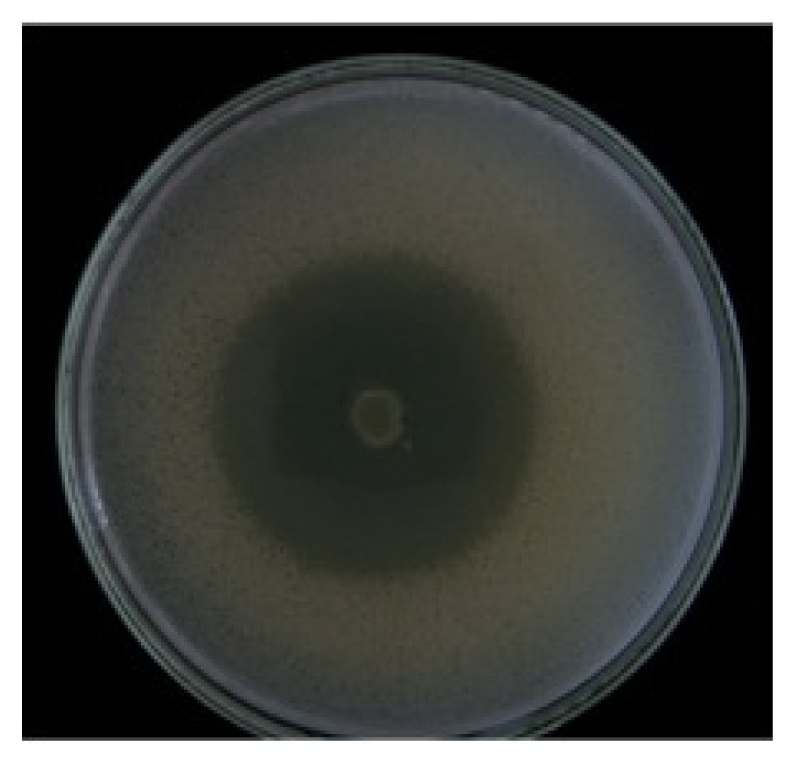
2. Results
3. Discussion
4. Experimental Section
4.1. Origin of Bacterial Strains
4.2. Molecular Characterization of the Environmental Strain
4.3. Inhibition Tests
4.4. Growth of the Environmental Strain and Extraction of Its Bioactive Products
4.5. Purification and Characterization of the Oleic Acid
4.6. Statistical Analysis
5. Conclusions
Acknowledgements
- Samples Availability: Available from the authors.
References
- Duque, B. Busqueda de compuestos bioactivos a partir de organismos marinos del caribe colombiano. Rev. Acad. Colomb. Cienc 1998, 22, 527–537. [Google Scholar]
- Newman, DJ; Cragg, GM. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod 2004, 67, 1216–1238. [Google Scholar]
- AL-Zereini, W. Natural Products from Marine Bacteria. Ph.D. Thesis, Technischen Universität Kaiserslautern, Kaiserslautern, Germany, September 2006. [Google Scholar]
- Márquez, D; Galeano, E; Martínez, A. Productos naturales con actividad antimicrobiana. Parte II. Vitae Rev. Fac. Quím. Farm 2004, 11, 35–41. [Google Scholar]
- Blunt, JW; Copp, BR; Munro, MHG; Northcote, PT; Prinsep, MR. Marine natural products. Nat. Prod. Rep 2006, 23, 26–78. [Google Scholar]
- Zapata, M; Silva, S; Luza, Y; Wilkens, M; Riquelme, C. The inhibitory effect of biofilms produced by wild bacterial isolates to the larval settlement of the fouling ascidia Ciona intestinalis and Pyura praeputialis. Electron. J. Biotechnol 2007, 10, 149–159. [Google Scholar]
- Zhang, Y; Mu, J; Gu, X; Zhao, C; Wang, X; Xie, Z. A Marine sulfate-reducing bacterium producing multiple antibiotics: Biological and chemical investigation. Mar. Drugs 2009, 7, 341–354. [Google Scholar]
- Parés, R; Juárez, A. Bioquímica de los Microorganismos; Editorial Reverté: Barcelona, Spain, 1997; pp. 1–380. [Google Scholar]
- Cai, J; Li, J; Thompson, KD; Li, C; Han, H. Isolation and characterization of pathogenic Vibrio parahaemolyticus from diseased post-larvae of abalone Haliotis diversicolor supertexta. J. Basic Microbiol 2007, 47, 84–86. [Google Scholar]
- Balcazar, JL; Rojas-Luna, T; Cunningham, D. Effect of the addition of four potential probiotic strains on the survival of pacific white shrimp (Litopenaeus vannamei) following immersion challenge with Vibrio parahaemolyticus. J. Invert. Pathol 2007, 96, 147–150. [Google Scholar]
- Isnansetyo, A; Istiqomah, I; Muhtadi; Sinansari, S; Hernawan, RK; Triyanto; Widada, J. A potential bacterial biocontrol agent, strain S2V2 against pathogenic marine Vibrio in aquaculture. World J. Microbiol. Biotechnol 2009, 25, 1103–1113. [Google Scholar]
- Harth, E; Matsuda, L; Hernández, C; Rioseco, ML; Romero, J; González-Escalona, N; Martínez-Urtaza, J; Espejo, R. Epidemiology of Vibrio parahaemolyticus Outbreaks, Southern Chile. J. Infect. Dis 2009, 15, 163–168. [Google Scholar]
- Zheng, CJ; Yoo, JS; Lee, TG; Cho, HY; Kim, YH; Kim, WG. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett 2005, 579, 5157–5162. [Google Scholar]
- Lekogo, BM; Coroller, L; Mathot, AG; Mafart, P; Leguerinel, I. Modelling the influence of palmitic, palmitoleic, stearic and oleic acids on apparent heat resistance of spores of Bacillus cereus NTCC 11145 and Clostridium sporogenes Pasteur 79.3. Int. J. Food Microbiol 2010, 141, 242–247. [Google Scholar]
- Nieman, C. Influence of trace amounts of fatty acids on the growth of microorganisms. Bacteriol. Rev 1954, 18, 147–163. [Google Scholar]
- Singh, IP; Milligan, KE; Gerwick, WH. Tanikolide, a toxic and antifungal lactone from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod 1999, 62, 1333–1335. [Google Scholar]
- Leyton, Y; Riquelme, C. Marine Bacillus spp. Associated with the egg capsule of Concholepas concholepas (common name “loco”) have an inhibitory activity toward the pathogen Vibrio parahaemolyticus. Microb. Ecol 2010, 60, 599–605. [Google Scholar]
- Jorquera, M; Riquelme, C; Loyola, L; Muñoz, L. Production of bactericidal substance by a marine Vibrio isolated from cultures of the scallop Argopecten purpuratus. Aquac. Int 1999, 7, 433–448. [Google Scholar]
- Sun, CQ; O’Connor, CJ; Roberton, AM. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol 2003, 36, 9–17. [Google Scholar]
- Fuenzalida, L; Armijo, L; Zabala, B; Hernández, C; Rioseco, ML; Riquelme, C; Espejo, R. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol 2007, 117, 270–275. [Google Scholar]
- Seidel, V; Taylor, PW. In vitro activity of extracts and constituents of Pelagonium against rapidly growing mycobacteria. Int. J. Antimicrob. Agents 2004, 23, 613–619. [Google Scholar]
- Sun, CQ; O’Connor, CJ; Roberton, AM. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol 2003, 36, 9–17. [Google Scholar]
- Farrington, M; Brenwald, N; Haines, D; Walpole, E. Resistance to desiccation and skin fatty acids in outbreak Straits of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol 1992, 36, 56–60. [Google Scholar]
- Nicolosi, RJ; Woolfrey, B; Wilson, TA; Scollin, P; Handelman, G; Fisher, R. Decreased aortic early atherosclerosis and associated risk factors in hypercholesterolemic hamsters fed a high or mid oleic acid oil compared to a high-linoleic acid oil. J. Nutr. Biochem 2004, 15, 540–547. [Google Scholar]
- Ziboh, VA; Miller, CC; Cho, Y. Metabolism of polyunsaturated fatty acids by skin epidermal enzymes: Generation of anti-inflammatory and antiproliferative metabolites. Am. J. Clin. Nutr 2000, 71, 361S–366S. [Google Scholar]
- Lunde, CS; Hartouni, SR; Janc, JW; Mammen, M; Humphrey, PP; Benton, BM. Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob. Agents Chemother 2009, 53, 3375–3383. [Google Scholar]
- Huang, C; George, B; Ebersole, JL. Antimicrobial activity of n-6, n-7 and n-9 fatty acids and their esters for oral microorganisms. Arch. Oral Biol 2010, 55, 555–560. [Google Scholar]
- Cardoso, CR; Favoreto, S; Oliveira, LL; Vancima, JO; Barbana, GB; Ferraza, DB; Silva, JS. Oleic acid modulation of the immune response in wound healing: A new approach for skin repair. Immunobiology 2010, 216, 409–415. [Google Scholar]
- Riquelme, C; Araya, R; Vergara, N; Rojas, R; Guaita, M; Candia, M. Potential of probiotic strains in the culture of the Chilean scallop Argopecten purpuratus (Lamarck, 1819). Aquaculture 1997, 154, 17–26. [Google Scholar]
- Sambrook, J; Fritsch, EF; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory: New York, NY, USA, 1989. [Google Scholar]
- Brosius, J; Dull, TJ; Sleeter, DD; Noller, HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol 1981, 148, 107–127. [Google Scholar]
- Thompson, D; Higgins, G; Gibson, J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar]
- Dopazo, CP; Lemos, ML; Bolinches, J; Barja, JL; Toranzo, AE. Inhibitory activity of antibiotic-producing marine bacteria against fish pathogens. J. Appl. Bacteriol 1988, 65, 97–101. [Google Scholar]
- Avendaño-Herrera, R; Lody, M; Riquelme, CE. Production of inhibitory substances among bacterial biofilms on marine substrates. Rev. Biol. Mar. Oceanogr 2005, 40, 117–125. [Google Scholar]
- Gerhardt, P; Murray, RGE; Wood, WA; Krieg, NR. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994. [Google Scholar]
- Porter, K; Feig, Y. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr 1980, 25, 943–948. [Google Scholar]
- Zar, J. Biostatistical Analysis; Prentice-Hall: New York, NY, USA, 1994; Samples Availability: Available from the authors. [Google Scholar]

© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Leyton, Y.; Borquez, J.; Darias, J.; Cueto, M.; Díaz-Marrero, A.R.; Riquelme, C. Oleic Acid Produced by a Marine Vibrio spp. Acts as an Anti-Vibrio parahaemolyticus Agent. Mar. Drugs 2011, 9, 2155-2163. https://doi.org/10.3390/md9102155
Leyton Y, Borquez J, Darias J, Cueto M, Díaz-Marrero AR, Riquelme C. Oleic Acid Produced by a Marine Vibrio spp. Acts as an Anti-Vibrio parahaemolyticus Agent. Marine Drugs. 2011; 9(10):2155-2163. https://doi.org/10.3390/md9102155
Chicago/Turabian StyleLeyton, Yanett, Jorge Borquez, José Darias, Mercedes Cueto, Ana R. Díaz-Marrero, and Carlos Riquelme. 2011. "Oleic Acid Produced by a Marine Vibrio spp. Acts as an Anti-Vibrio parahaemolyticus Agent" Marine Drugs 9, no. 10: 2155-2163. https://doi.org/10.3390/md9102155



