Pardaxin, an Antimicrobial Peptide, Triggers Caspase-Dependent and ROS-Mediated Apoptosis in HT-1080 Cells
Abstract
:1. Introduction
2. Results
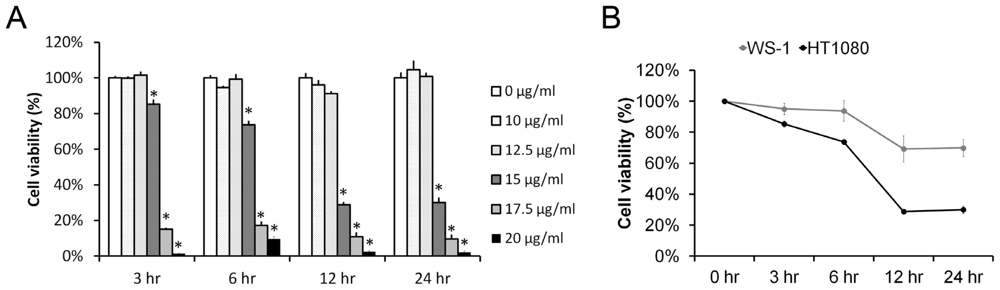
2.1. Pardaxin Inhibited Cell Proliferation in Human Fibrosarcoma HT-1080 Cells
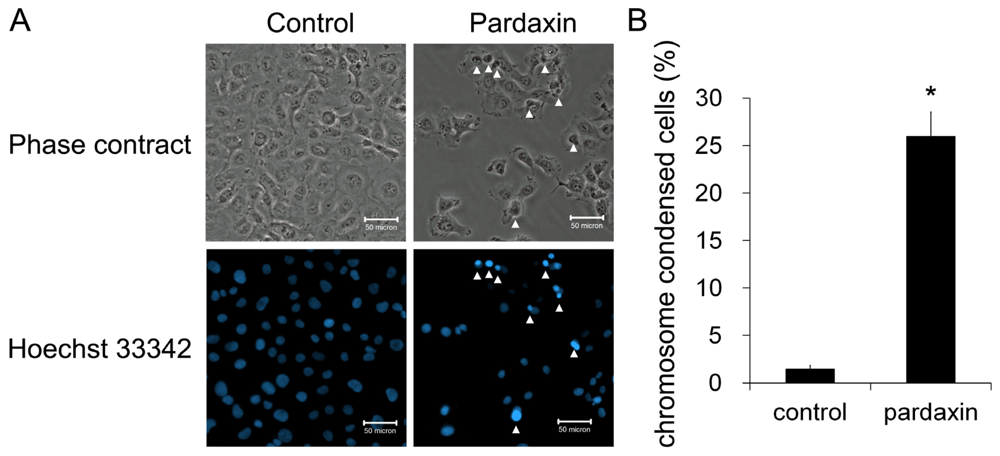
2.2. Pardaxin Induced Apoptosis in HT-1080 Cells
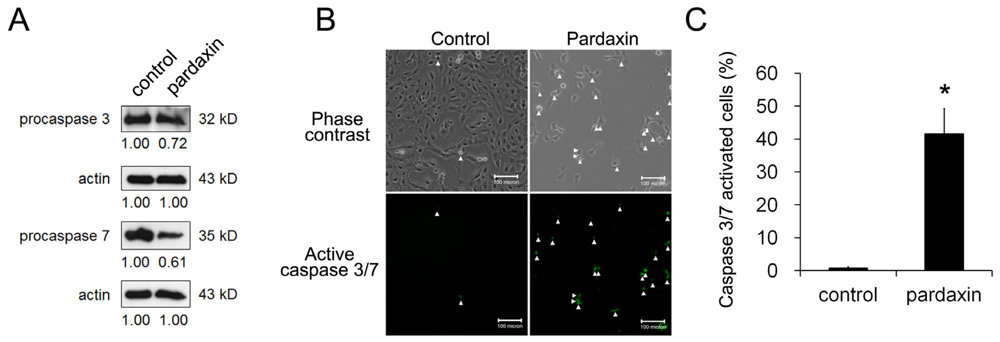
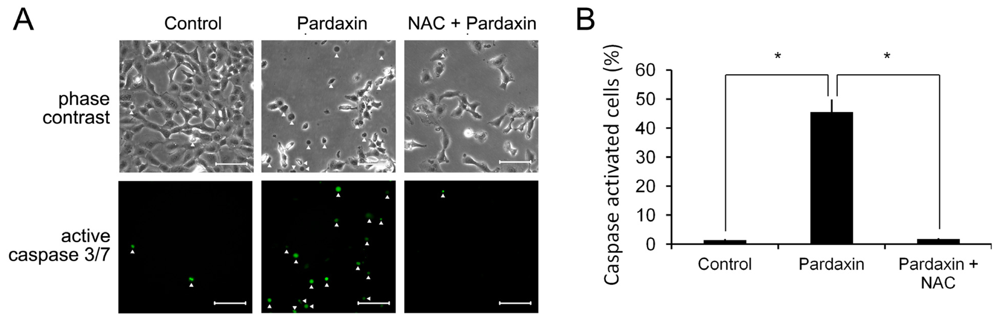
2.3. Pardaxin Activated Caspase-3/7 Activities
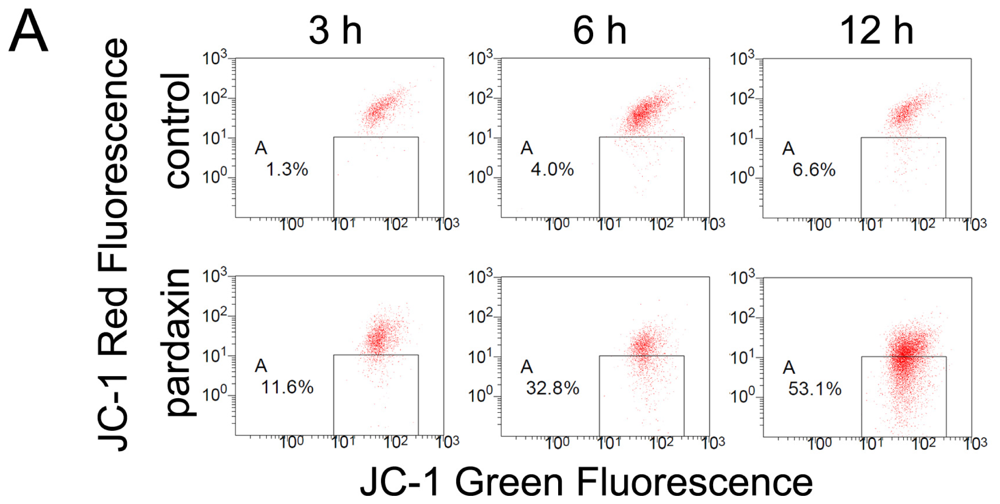
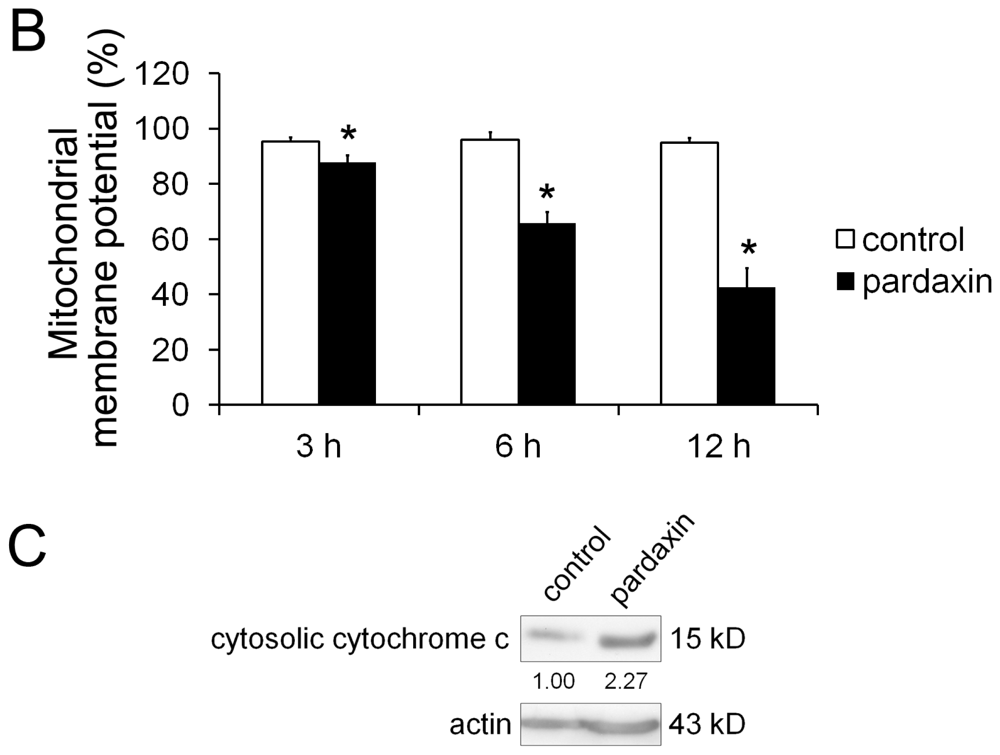
2.4. Pardaxin Caused the Loss of the Mitochondrial Membrane Potential (MMP) and the Release of Cyt c in HT-1080 Cells
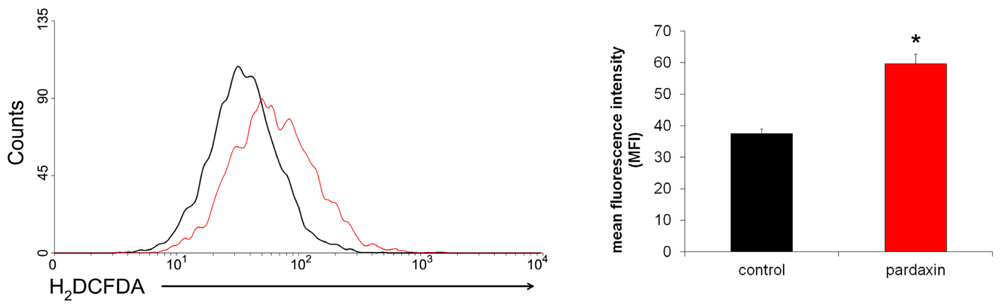
2.5. Pardaxin Increased the Production of ROS in HT-1080 Cells
2.6. Pardaxin Activation of Caspases-3/7 Is ROS Mediated
2.7. Pardaxin-Induced Cell Death Is Caspase-3/7 Dependent
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Annexin V/Propidium Iodide (PI) Staining
4.5. Hoechst 33342 Staining
4.6. Mitochondrial Membrane Potential (MMP) Detection
4.7. Measurement of Intracellular ROS
4.8. Caspase-3/7 Activity Assay
4.9. Western Blot Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Material
marinedrugs-09-01955-s001.pdfAcknowledgements
References
- Ellis, HM; Horvitz, HR. Genetic control of programmed cell death in the nematode C. elegans. Cell 1986, 44, 817–829. [Google Scholar]
- Hengartner, MO; Horvitz, HR. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 1994, 76, 665–676. [Google Scholar]
- Miura, M; Zhu, H; Rotello, R; Hartwieg, EA; Yuan, J. Induction of apoptosis in fibroblasts by IL-1 [beta]-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell 1993, 75, 653–660. [Google Scholar]
- Chinnaiyan, AM; O’Rourke, K; Lane, BR; Dixit, VM. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 1997, 275, 1122–1126. [Google Scholar]
- Riedl, SJ; Salvesen, GS. The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol 2007, 8, 405–413. [Google Scholar]
- Boatright, KM; Salvesen, GS. Mechanisms of caspase activation. Curr. Opin. Cell Biol 2003, 15, 725–731. [Google Scholar]
- Hockenbery, DM; Oltvai, ZN; Yin, XM; Milliman, CL; Korsmeyer, SJ. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell 1993, 75, 241–251. [Google Scholar]
- Gross, A; McDonnell, JM; Korsmeyer, SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev 1999, 13, 1899–1911. [Google Scholar]
- Buttke, TM; Sandstrom, PA. Oxidative stress as a mediator of apoptosis. Immunol. Today 1994, 15, 7–10. [Google Scholar]
- Zou, H; Li, Y; Liu, X; Wang, X. An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem 1999, 274, 11549–11556. [Google Scholar]
- Chang, HY; Yang, X. Proteases for cell suicide: functions and regulation of caspases. Microbiol. Mol. Biol. Rev 2000, 64, 821–846. [Google Scholar]
- Budihardjo, I; Oliver, H; Lutter, M; Luo, X; Wang, X. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol 1999, 15, 269–290. [Google Scholar]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol 2007, 35, 495–516. [Google Scholar]
- Kerr, JFR; Winterford, CM; Harmon, BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer 1994, 73, 2013–2026. [Google Scholar]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar]
- Tossi, A; Sandri, L. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr. Pharm. Des 2002, 8, 743–761. [Google Scholar]
- Epand, RM; Vogel, HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1999, 1462, 11–28. [Google Scholar]
- Brogden, KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol 2005, 3, 238–250. [Google Scholar]
- Mader, JS; Hoskin, DW. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs 2006, 15, 933–946. [Google Scholar]
- Kim, YJ; Varki, A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj. J 1997, 14, 569–576. [Google Scholar]
- Clark, E; Chao, S. A toxic secretion from the Red Sea flatfish Pardachirus marmoratus(Lacepede). Sea Fish. Res. Sta. Haifa Bull 1973, 60, 53–56. [Google Scholar]
- Shai, Y; Fox, J; Caratsch, C; Shih, YL; Edwards, C; Lazarovici, P. Sequencing and synthesis of pardaxin, a polypeptide from the Red Sea Moses sole with ionophore activity. FEBS Lett 1988, 242, 161–166. [Google Scholar]
- Lelkes, PI; Lazarovici, P. Pardaxin induces aggregation but not fusion of phosphatidylserine vesicles. FEBS Lett 1988, 230, 131–136. [Google Scholar]
- Pal, R; Barenholz, Y; Wagner, RR. Pardaxin, a hydrophobic toxin of the Red Sea flatfish, disassembles the intact membrane of vesicular stomatitis virus. J. Biol. Chem 1981, 256, 10209–10212. [Google Scholar]
- Thennarasu, S; Nagaraj, R. Specific antimicrobial and hemolytic activities of 18-residue peptides derived from the amino terminal region of the toxin pardaxin. Protein Eng 1996, 9, 1219–1224. [Google Scholar]
- Hsu, JC; Lin, LC; Tzen, JTC; Chen, JY. Pardaxin-induced apoptosis enhances antitumor activity in HeLa cells. Peptides 2011, 32, 1110–1116. [Google Scholar]
- Orrenius, S. Reactive oxygen species in mitochondria-mediated cell death. Drug Metab. Rev 2007, 39, 443–455. [Google Scholar]
- Li, PF; Dietz, R; von Harsdorf, R. P53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J 1999, 18, 6027–6036. [Google Scholar]
- Andreu, D; Rivas, L. Animal antimicrobial peptides: An overview. Biopolymers 1998, 47, 415–434. [Google Scholar]
- Matsuzaki, K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1999, 1462, 1–10. [Google Scholar]
- Brugger, B; Erben, G; Sandhoff, R; Wieland, F; Lehmann, W. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1997, 94, 2339–2344. [Google Scholar]
- van Meer, G; Voelker, DR; Feigenson, GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 2008, 9, 112–124. [Google Scholar]
- Kavanagh, K; Dowd, S. Histatins: Antimicrobial peptides with therapeutic potential. J. Pharm. Pharmacol 2004, 56, 285–289. [Google Scholar]
- Zughaier, SM; Shafer, WM; Stephens, DS. Antimicrobial peptides and endotoxin inhibit cytokine and nitric oxide release but amplify respiratory burst response in human and murine macrophages. Cell Microbiol 2005, 7, 1251–1262. [Google Scholar]
- Bloch-Shilderman, E; Jiang, H; Lazarovici, P. Pardaxin, an ionophore neurotoxin, induces PC12 cell death: activation of stress kinases and production of reactive oxygen species. J. Nat. Toxins 2002, 11, 71–85. [Google Scholar]
- Matés, JM; Segura, JA; Alonso, FJ; Márquez, J. Intracellular redox status and oxidative stress: Implications for cell proliferation, apoptosis, and carcinogenesis. Arch. Toxicol 2008, 82, 273–299. [Google Scholar]
- Armstrong, JS; Jones, DP. Glutathione depletion enforces the mitochondrial permeability transition and causes cell death in Bcl-2 overexpressing HL60 cells. FASEB J 2002, 16, 1263–1265. [Google Scholar]
- Reed, E. Platinum-DNA adduct, nucleotide excision repair and platinum based anti-cancer chemotherapy. Cancer Treat. Rev 1998, 24, 331–344. [Google Scholar]


© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Huang, T.-C.; Lee, J.-F.; Chen, J.-Y. Pardaxin, an Antimicrobial Peptide, Triggers Caspase-Dependent and ROS-Mediated Apoptosis in HT-1080 Cells. Mar. Drugs 2011, 9, 1995-2009. https://doi.org/10.3390/md9101995
Huang T-C, Lee J-F, Chen J-Y. Pardaxin, an Antimicrobial Peptide, Triggers Caspase-Dependent and ROS-Mediated Apoptosis in HT-1080 Cells. Marine Drugs. 2011; 9(10):1995-2009. https://doi.org/10.3390/md9101995
Chicago/Turabian StyleHuang, Tsui-Chin, Jheng-Fong Lee, and Jyh-Yih Chen. 2011. "Pardaxin, an Antimicrobial Peptide, Triggers Caspase-Dependent and ROS-Mediated Apoptosis in HT-1080 Cells" Marine Drugs 9, no. 10: 1995-2009. https://doi.org/10.3390/md9101995



