Levantilides A and B, 20-Membered Macrolides from a Micromonospora Strain Isolated from the Mediterranean Deep Sea Sediment
Abstract
:1. Introduction
2. Results and Discussion

3. Experimental Section
3.1. Isolation and identification of strain M71-A77
3.2. Chemical analysis
General experimental procedures
Isolation of levantilides A and B
3.3. Production of levantilide A (1) and B (2) at in situ salinity
3.4. Antimicrobial tests
3.5. Cytotoxic tests
Acknowledgement
- Supplementary Information1H NMR spectra, 13C NMR spectra, COSY spectra and HMBC spectra of compounds 1 and 2 as well as the NMR spectroscopic data of compound 2 are available as supplementary information.
References
- Lam, KS. Discovery of novel metabolites from marine actinomycetes. Curr Opin Microbiol 2006, 9, 245–251. [Google Scholar]
- Berdy, J. Bioactive microbial metabolites. J Antibiot 2005, 58, 1–26. [Google Scholar]
- Jensen, PR; Mincer, TJ; Williams, PG; Fenical, W. Marine actinomycete diversity and natural product discovery. Antonie van Leeuwenhoek 2005, 87, 43–48. [Google Scholar]
- Fiedler, HP; Bruntner, C; Bull, AT; Ward, AC; Goodfellow, M; Potterat, O; Puder, C; Mihm, G. Marine actinomycetes as a source of novel secondary metabolites. Antonie van Leeuwenhoek 2005, 87, 37–42. [Google Scholar]
- Magarvey, NA; Keller, JM; Bernan, V; Dworkin, M; Sherman, DH. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl Environ Microbiol 2004, 70, 7520–7529. [Google Scholar]
- Bernan, VS; Greenstein, M; Carter, GT. Mining marine microorganisms as a source of new antimicrobials and antifungals. Curr Med Chem-Anti-Infect Agents 2004, 3, 181–195. [Google Scholar]
- Hohmann, C; Schneider, K; Bruntner, C; Irran, E; Nicholson, G; Bull, AT; Jones, AL; Brown, R; Stach, JEM; Goodfellow, M; et al. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937*. J Antibiot 2009, 62, 99–104. [Google Scholar]
- Hohmann, C; Schneider, K; Bruntner, C; Brown, R; Jones, AL; Goodfellow, M; Kramer, M; Imhoff, JF; Nicholson, G; Fiedler, HP; et al. Albidopyrone, a new [alpha]-pyrone-containing metabolite from marine-derived Streptomyces sp. NTK 227*. J Antibiot 2009, 62, 75–79. [Google Scholar]
- Thingstad, TF; Krom, MD; Mantoura, RFC; Flaten, GAF; Groom, S; Herut, B; Kress, N; Law, CS; Pasternak, A; Pitta, P. Nature of phosphorus limitation in the ultraoligotrophic Eastern Mediterranean. Science 2005, 309, 1068–1071. [Google Scholar]
- Katz, L; Ashley, GW. Translation and protein synthesis: macrolides. Chem Rev 2005, 105, 499–528. [Google Scholar]
- Ohta, E; Ohta, S; Kubota, NK; Suzuki, M; Ogawa, T; Yamasaki, A; Ikegami, S. Micromonospolide A, a new macrolide from Micromonospora sp. Tetrahedron Lett 2001, 42, 4179–4181. [Google Scholar]
- Weinstein, MJ; Wagman, GH; Marquez, JA; Testa, RT; Oden, E; Waitz, JA. Megalomicin, a new macrolide antibiotic complex produced by Micromonospora. J Antibiot 1969, 22, 253–258. [Google Scholar]
- Hatano, K; Higashide, E; Shibata, M. Studies on juvenimicin, a new antibiotic I. J Antibiot 1976, 29, 1163–1170. [Google Scholar]
- Satoi, S; Muto, N; Hayashi, M; Fujii, T; Otani, M. Mycinamicins, new macrolide antibiotics. I. J Antibiot 1980, 33, 364–376. [Google Scholar]
- Waitz, JA; Drube, CG; Moss, EL; Weinstein, MJ. Biological studies with rosamicin, a new micomonospora-produced macrolide antibiotic. J Antibiot 1972, 25, 647–652. [Google Scholar]
- Kobayashi, J; Ishibashi, M; Nakamura, H; Ohizumi Terufumi, Y. Amphidinolide-A, a novel antineoplastic macrolide from the marine dinoflagellate sp. Tetrahedron Lett 1986, 27, 5755–5758. [Google Scholar]
- Tsuda, M; Endo, T; Kobayashi, J. Amphidinolide U, novel 20-membered macrolide from marine dinoflagellate amphidinium sp. Tetrahedron 1999, 55, 14565–14570. [Google Scholar]
- Tsuda, M; Oguchi, K; Iwamoto, R; Okamoto, Y; Fukushi, E; Kawabata, J; Ozawa, T; Masuda, A; Kitaya, Y; Omasa, K. Iriomoteolide-1a, a potent cytotoxic 20-membered macrolide from a benthic dinoflagellate Amphidinium species. J Org Chem 2007, 72, 4469–4474. [Google Scholar]
- Tsuda, M; Oguchi, K; Iwamoto, R; Okamoto, Y; Fukushi, E; Kawabata, J; Ozawa, T; Masuda, A. Iriomoteolides-1b and -1c, 20-membered macrolides from a marine dinoflagellate Amphidinium species. J Nat Prod 2007, 70, 1661–1663. [Google Scholar]
- Kobayashi, J; Tsuda, M. Amphidinolides, bioactive macrolides from symbiotic marine dinoflagellates. Nat Prod Rep 2004, 21, 77–93. [Google Scholar]
- Kobayashi, J; Ishibashi, M. Bioactive metabolites of symbiotic marine microorganisms. Chem Rev 1993, 93, 1753–1769. [Google Scholar]
- Kobayashi, J; Kubota, T. Bioactive macrolides and polyketides from marine dinoflagellates of the genus Amphidinium. J Nat Prod 2007, 70, 451–460. [Google Scholar]
- Skropeta, D. Deep-sea natural products. Nat Prod Rep 2008. [Google Scholar]
- Pathom-aree, W; Stach, J; Ward, A; Horikoshi, K; Bull, A; Goodfellow, M. Diversity of actinomycetes isolated from Challenger Deep sediment (10.898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar]
- Prieto-Davo, A; Fenical, W; Jensen, PR. Comparative actinomycete diversity in marine sediments. Aquat Microb Ecol 2008, 52, 1–11. [Google Scholar]
- Colquhoun, JA; Heald, SC; Li, L; Tamaoka, J; Kato, C; Horikoshi, K; Bull, AT. Taxonomy and biotransformation activities of some deep-sea actinomycetes. Extremophiles 1998, 2, 269–277. [Google Scholar]
- Gärtner, A; Wiese, J; Imhoff, JF. Amphritea atlantica gen. nov., sp. nov., a gammaproteobacterium from the Logatchev hydrothermal vent field. Int J Syst Evol Microbiol 2008, 58, 34–39. [Google Scholar]
- Lang, G; Wiese, J; Schmaljohann, R; Imhoff, JF. New pentaenes from the sponge-derived marine fungus Penicillium rugulosum: structure determination and biosynthetic studies. Tetrahedron 2007, 63, 11844–11849. [Google Scholar]
- Dengler, WA; Schulte, J; Berger, DP; Mertelsmann, R; Fiebig, HH. Development of a propidium iodide fluorescence assay for proliferation and cytotoxicity assays. Anti-Cancer Drugs 1995, 6. [Google Scholar]
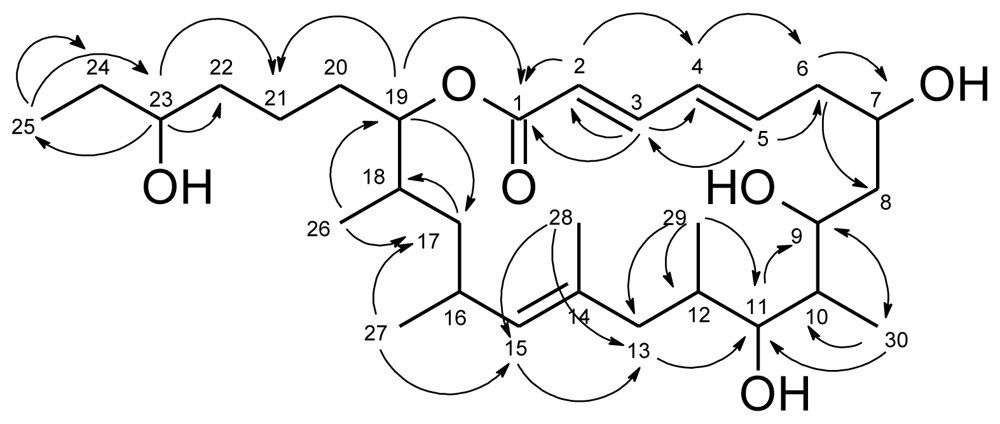
| levantilide A (1) | |||||
|---|---|---|---|---|---|
| position | δC | δH, J [Hz] | COSY | HMBC | NOESY |
| 1 | 166.1, C | ||||
| 2 | 120.4, CH | 5.78, d (15.5) | 3 | 1, 3, 4, 5 | 3, 4 |
| 3 | 143.8, CH | 7.06, dd (11.2, 15.5) | 2, 4 | 1, 2, 4, 5 | 2, 4, 5 |
| 4 | 130.6, CH | 6.29, dd (11.2, 15.2) | 3, 5 | 2, 3, 6 | 2, 3, 5, 6b |
| 5 | 139.8, CH | 6.08, ddd (4.4, 10, 15.2) | 4, 6 | 3, 6, 7 | 3, 4, 6a, 7 |
| 6a | 39.6, CH2 | 2.57, m | 5, 6b, 7 | 4, 5, 7, 8 | 5 |
| 6b | 2.33, dt (14.4, 9.9) | 5, 6a, 7 | 4, 5, 7, 8 | 4 | |
| 7 | 67.2, CH | 3.98, m | 6, 7-OH, 8 | 8b, 5 | |
| 7-OH | 4.81, br. d (3.8) | 7 | 7, 8 | ||
| 8a | 33.4, CH2 | 1.51a, m | 7, 8b, 9 | 6, 7, 9 | 8b |
| 8b | 1.16, ddd (14.6, 5.6, 2.3) | 7, 8a, 9 | 6, 7, 9 | 7, 8a, 11 | |
| 9 | 67.4, CH | 3.92, br. d (11.2) | 8, 9-OH, 10 | 30 | 12 |
| 9-OH | 4.63, br. s | 9 | 8, 9, 10 | ||
| 10 | 41.0, CH | 1.68, m | 9, 11, 30 | ||
| 11 | 75.3, CH | 2.98 br. ddd (8.9, 6.0, 1.8) | 10, 11-OH, 12 | 9, 10, 12, 13, 29, 30 | 8b, 29 |
| 11-OH | 4.00, d (6.0) | 11 | 10, 11, 12 | ||
| 12 | 32.1, CH | 1.24, m | 11, 13, 29 | 9 | |
| 13a | 40.1, CH2 | 1.87, br. d (12.8) | 12, 13b, 15, 29 | 11, 12, 14, 15, 27, 28, 29 | 13b |
| 13b | 1.51a, m | 12, 13a, 15, 29 | 11, 12, 14, 15, 27, 28, 29 | 13a, 15 | |
| 14 | 132.8, C | ||||
| 15 | 132.7, CH | 4.76, d (7.8) | 13, 16 | 12, 13, 16, 17, 27, 28 | 13b, 17, 29 |
| 16 | 29.4, CH | 2.59, m | 15, 17, 27 | 14, 17, 27 | 27, 28 |
| 17a | 40.9, CH2 | 1.34, m | 16, 17b, 18 | 15, 16, 18, 19, 26, 27 | 17b |
| 17b | 1.05, ddd (13.5, 8.7, 5.0) | 16, 17a, 18 | 15, 16, 18, 19, 26, 27 | 17a, 27 | |
| 18 | 33.4, CH | 1.75, m | 17, 19, 26 | 16, 17, 19, 20, 26 | |
| 19 | 77.4, CH | 4.71, dt (10, 2.4) | 18, 20 | 1, 17, 20, 21,26 | 21, 26 |
| 20a | 27.9, CH2 | 1.52a, m | 19, 21 | ||
| 20b | 1.47a, m | 19, 21 | |||
| 21 | 22.0, CH2 | 1.30b, m | 20 | 19 | |
| 22 | 36.1, CH2 | 1.31b, m | 23 | 23, 23-OH | |
| 23 | 70.8, CH | 3.28, m | 22, 23-OH, 24 | 21, 22, 25 | 22, 25, 26 |
| 23-OH | 4.25, d (5.2) | 23 | 22, 23, 24 | 22, 26 | |
| 24a | 29.7, CH2 | 1.30b, m | 23, 25 | 25 | |
| 24b | 1.27b, m | 23, 25 | 25 | ||
| 25 | 10.1, CH3 | 0.82, t (7.4) | 24 | 23, 24 | 23, 24 |
| 26 | 17.5, CH3 | 0.88, d (6.2) | 18 | 17, 18, 19 | 19, 23, 23-OH |
| 27 | 21.5, CH3 | 0.84, d (6.8) | 16 | 15, 16, 17 | 16, 17b |
| 28 | 17.0, CH3 | 1.53a, s | 13, 14, 15 | 16 | |
| 29 | 17.7, CH3 | 0.58, d (6.7) | 12 | 11, 12, 13 | 11, 15 |
| 30 | 11.1, CH3 | 0.86, d (5.9) | 10 | 9, 10, 11 | 9, 11 |
| levantilide A (1) | levantilide B (2) | |||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 166.9 | 166.9 | ||
| 2 | 122.0 | 5.83 | 121.9 | 5.83 |
| 3 | 144.3 | 7.14 | 144.4 | 7.15 |
| 4 | 131.9 | 6.36 | 131.9 | 6.36 |
| 5 | 140.1 | 6.09 | 140.2 | 6.11 |
| 6a | 40.6 | 2.69 | 40.6 | 2.66 |
| 6b | 2.47 | 2.46 | ||
| 7 | 69.3 | 4.11 | 69.4 | 4.10 |
| 8a | 33.8 | 1.73 | 33.9 | 1.72 |
| 8b | 1.34 | 1.34 | ||
| 9 | 69.3 | 4.18 | 69.4 | 4.17 |
| 10 | 42.2 | 1.86 | 42.3 | 1.86 |
| 11 | 77.4 | 3.17 | 77.4 | 3.18 |
| 12 | 33.2 | 1.41 | 33.3 | 1.41 |
| 13a | 41.4 | 2.00 | 41.4 | 1.99 |
| 13b | 1.69 | 1.69 | ||
| 14 | 134.1 | 134.1 | ||
| 15 | 134.1 | 4.86 | 134.1 | 4.87 |
| 16 | 30.7 | 2.70 | 30.7 | 2.69 |
| 17a | 41.9 | 1.47 | 41.9 | 1.43 |
| 17b | 1.12 | 1.11 | ||
| 18 | 34.7 | 1.87 | 34.7 | 1.87 |
| 19 | 78.8 | 4.83 | 78.6 | 4.81 |
| 20a | 28.9 | 1.61 | 28.3 | 1.57 |
| 20b | 1.53 | 1.50 | ||
| 21a | 23.3 | 1.42 | 21.3 | 1.62 |
| 21b | 1.47 | |||
| 22 | 37.6 | 1.42 | 42.1 | 2.45 |
| 23 | 72.7 | 3.42 | 210.5 | |
| 24a | 31.1 | 1.43 | 36.0 | 2.42 |
| 24b | 1.37 | |||
| 25 | 10.4 | 0.90 | 8.0 | 0.96 |
| 26 | 18.6 | 0.91 | 18.4 | 0.91 |
| 27 | 22.0 | 0.86 | 21.8 | 0.87 |
| 28 | 17.6 | 1.62 | 17.6 | 1.63 |
| 29 | 18.5 | 0.70 | 18.4 | 0.71 |
| 30 | 10.8 | 0.98 | 11.0 | 0.98 |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gärtner, A.; Ohlendorf, B.; Schulz, D.; Zinecker, H.; Wiese, J.; Imhoff, J.F. Levantilides A and B, 20-Membered Macrolides from a Micromonospora Strain Isolated from the Mediterranean Deep Sea Sediment. Mar. Drugs 2011, 9, 98-108. https://doi.org/10.3390/md9010098
Gärtner A, Ohlendorf B, Schulz D, Zinecker H, Wiese J, Imhoff JF. Levantilides A and B, 20-Membered Macrolides from a Micromonospora Strain Isolated from the Mediterranean Deep Sea Sediment. Marine Drugs. 2011; 9(1):98-108. https://doi.org/10.3390/md9010098
Chicago/Turabian StyleGärtner, Andrea, Birgit Ohlendorf, Dirk Schulz, Heidi Zinecker, Jutta Wiese, and Johannes F. Imhoff. 2011. "Levantilides A and B, 20-Membered Macrolides from a Micromonospora Strain Isolated from the Mediterranean Deep Sea Sediment" Marine Drugs 9, no. 1: 98-108. https://doi.org/10.3390/md9010098




