1. Introduction
The deep sea is characterized by low temperature (1–4 °C), extremely high hydrostatic pressure, lack of sunlight, and relatively low influx of utilizable organic material derived from primary production in surface waters. Among such environmental factors, hydrostatic pressure is thought to have the greatest influence on the vertical distribution of organisms and speciation in the deep seas [
1–
3] and on the formation of protein complexes, e.g., enzyme–substrate or protein–protein interactions [
4]. Many previous studies reported proteins from deep-sea fishes that function at high pressure [
4–
6], and hypothetical models of protein adaptation to deep-sea pressure have been proposed [
4]. However, the primary structures of those proteins have not been determined in detail [
3]. The effects of pressure on the K
m values of many enzymes from shallow- and deep-dwelling marine invertebrates and fishes were studied [
5]. Malate dehydrogenase from organisms adapted to pressure of 0.1 MPa increased with additional pressure, resulting in a concomitant increase in the K
m values for nicotinamide adenine dinucleotide (NADH) [
7]. The α-skeletal actin cDNA from
Coryphaenoides sp. was cloned and sequenced, and the specific amino acid substitutions responsible for the adaptation of α-actin to high pressure were elucidated [
3]. The kinetic properties and amino acid sequences of lactate dehydrogenase (LDH) expressed in skeletal muscle [L-LDH: NAD oxidoreductase, EC 1.1.1.27; (LDH-A
4)] were compared among orthologs from congeners of Pacific damselfish, and temperature-adaptive changes in LDH-A
4 structure and function were found [
8]. When the kinetic properties and amino acid sequences of LDH in heart muscle (LDH-B
4) from two cod species were compared, pressure-adaptive changes in LDH-B
4 structure and function were observed. The results of circular dichroism spectroscopic analysis pointed to protein unfolding as the cause of inactivation [
9]. In those studies, however, the amino acids responsible for the adaptation were not identified.
We previously reported the kinetic properties and amino acid sequences of LDH-A
4 from three hagfishes inhabiting different depths [
10]. The activity of LDH-A from the shallow-sea hagfish
Eptatretus burgeri was almost completely lost when subjected to pressure of 50 MPa for three minutes, while that from the deep-sea hagfish
Eptatretus okinoseanus did not change when subjected to the same conditions, and thus a pressure-dependent change in LDH-A
4 structure was proposed [
11]. In this study, we investigated the pressure-adaptive mechanism of LDHs from hagfish and considered the relationship between this mechanism and human disease.
2. Results and Discussion
The phylogenies of hagfish and lampreys in the Cyclostomata are not yet clear-cut. In this study, we compared their isozymes using electrophoresis.
Figure 1 shows the electrophoretic patterns of LDH isozymes from the skeletal muscle and heart of two hagfishes,
E. okinoseanus and
Paramyxine atami, and a lamprey,
Entosphenus japonica. Lampreys are also jawless fish, and their evolutionary relationship with hagfish is under debate. The skeletal muscle of the three species had the A
4 isozyme (
Figure 1, lanes 2, 4, and 6). The B
4 isozyme was expressed in the hearts of the two hagfish (lanes 3 and 5), but not in the lamprey (lane 1). Whereas human serum LDH had heteroisozymes (lane 7), such as A
1B
3, A
2B
2, and A
3B
1, hagfish LDHs did not.
Figure 1 indicates that hagfish diverged after lampreys in the evolution of vertebrates.
As shown in
Figure 2, the thermal stability of LDH activity was examined in the reaction from pyruvic acid to lactic acid at temperatures ranging from 10 to 60 °C during 30-min incubation. At 60 °C, the activity of LDH
from E. okinoseanus and E. burgeri decreased to about 40% of the original value.
The activity of
E. burgeri remained constant with heat treatment in the range from 10 °C to 55 °C, whereas that of
E. okinoseanus gradually decreased. Thus, the heat stability of LDH-A
4 from the hagfish differs depending on the depth of their habitats. The reason why LDHs from the hagfish show these thermophilic properties is not yet clear. Johns and Somero reported that the heat stability of Pacific damselfish LDH was achieved by amino acid substitution of residue 219 from Thr to Ala [
8]. In the three hagfish LDHs examined in this study, amino acid 219 was Leu and not relevant to the differences in heat stability among them. None of the additional region that is specific and common to hagfish LDHs (amino acids 220–227), is responsible for heat stability.
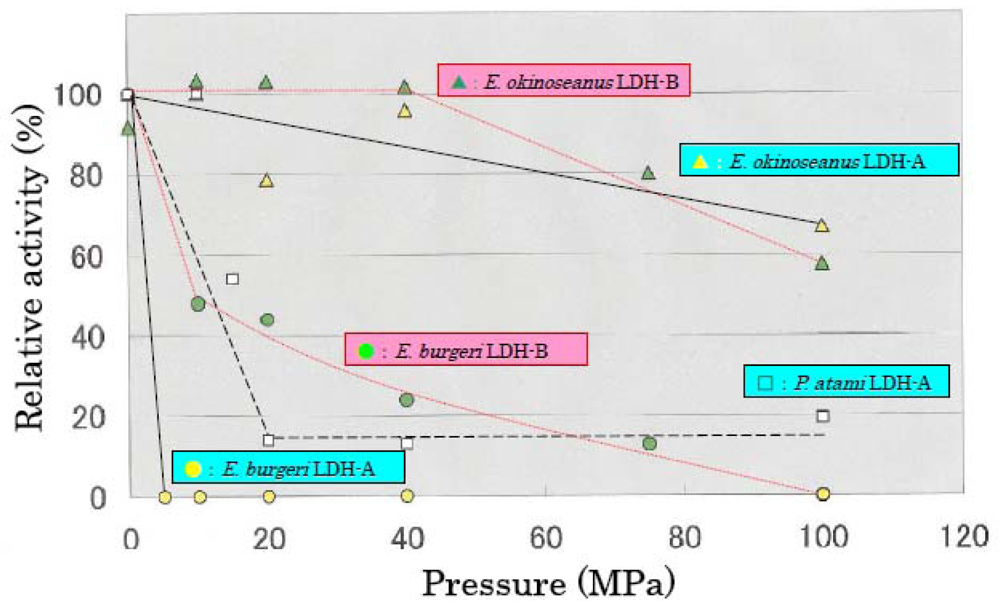
The effects of high hydrostatic pressure from 0.1–100 MPa on LDH activities from the three hagfishes were examined using high-pressure photometry at the Japan Agency for Marine-Earth Science and Technology.
Figure 3 shows the effects of hydrostatic pressure on LDH activities. LDH-A and -B from
E. okinoseanus, living at a depth of 1000 m, were highly active at high pressure of 100 MPa, maintaining 70% of that observed at 0.1 MPa. In contrast, the activities of LDH-A and -B from
P. atami, living at 250–400 m, decreased to 55% at 15 MPa, and those from
E. burgeri, living at 45–60 m, were completely lost at 5 MPa. These results show that the deeper the habitat, the greater the tolerance to pressure.
We compared the electrophoretic patterns of the LDH-As from
E. okinoseanus and
E. burgeri under high-pressure conditions. The tetrameric structure of LDH-As from
E. okinoseanus did not change at 50 MPa. In contrast, almost all LDH tetramers from
E. burgeri dissociated to dimmers and monomers at 50 MPa but reverted to tetramers at 0.1 MPa. These results show that the dissociation of tetramers caused the inactivation of
E. burgeri LDH. The mechanism of the slight gradual inactivation of
E. okinoseanus LDH at high pressure differs and is probably due to the metamorphosis of its inner structures [
12]. The results will be published elsewhere.
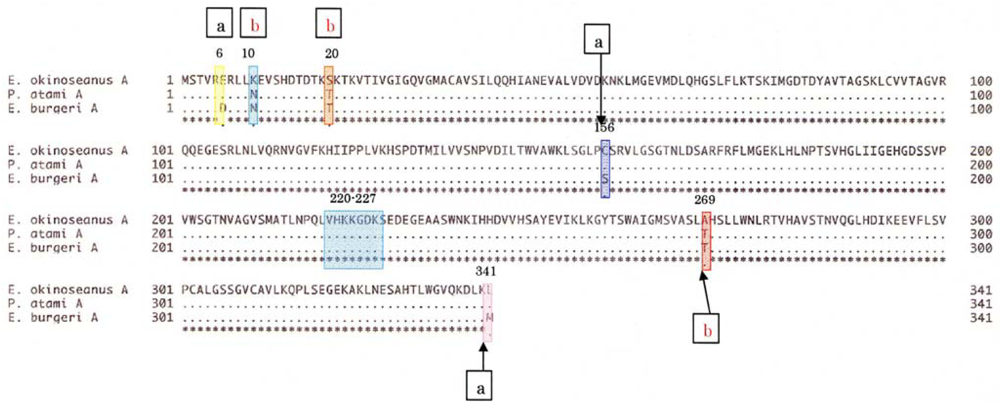
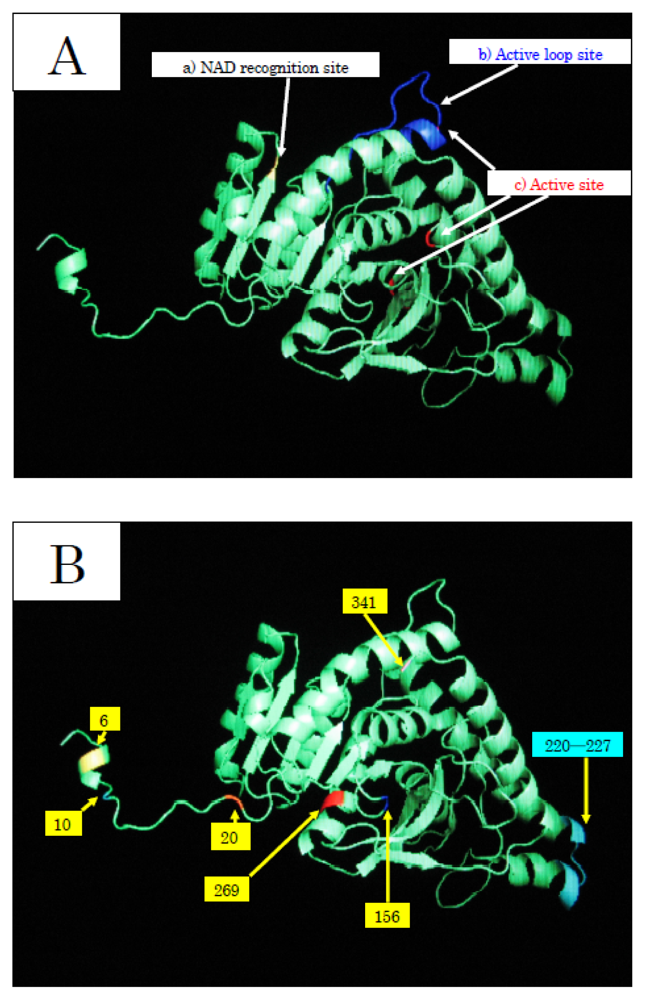
To elucidate the molecular mechanisms of the adaptation to high pressure, we compared the amino acid sequences (
Figure 4) and three-dimensional structures (
Figure 5) of LDHs in these hagfish. There were differences in six amino acid residues (6, 10, 20, 156, 269, and 341) in LDHs of the hagfishes, and additional regions specific to hagfish LDHs were found.
Two of the amino acids (156 and 269) are in the neighborhood of the active sites, and thus may control enzymatic activity. The other four amino acids (6, 10, 20, and 341) may be assigned to the part that combines four monomers into a tetramer.
The differences in the amino acid at position of the hagfish LDHs could affect their pressure resistance. Those amino acids, Asp in
E. burgeri LDH-A
4 and Glu in
E. okinoseanus LDH-A
4, have isoelectric points of 2.77 and 3.22, respectively. The deeper the habitat of the hagfish, the weaker the negative charge of the 6th amino acid. The core structures of hagfish LDHs inhabiting deeper water are only slightly affected by water. The differences in the 10th amino acid of the hagfish LDHs could affect their pressure resistance. Those amino acids are Asn in
E. burgeri LDH-A
4 and Lys in
E. okinoseanus LDH-A
4. The Lys of
E. okinoseanus LDH-A
4 is more hydrophilic than the Asn in
E. burgeri LDH-A
4, and thus the combination of four monomers into a tetramer in
E. okinoseanus LDH-A
4 may be tight. The N-terminal 20 residues that extend from the main body of the subunit are important in the interaction between subunits [
13]. As the other amino acids (20 and 341) that are also located at the binding position in the tetramer have almost the same properties, they may not affect pressure resistance. The differences in the 269th amino acid residue of the hagfish LDHs, located in part of the core structure, could be related to the tolerance to hydrostatic pressure. The amino acids Thr in
E. burgeri LDH-A
4 and Ala in
E. okinoseanus LDH-A
4 have molecular weights of 119 and 89, respectively. The core structure of
E. okinoseanus LDH is barely affected by hydrostatic pressure, because Ala is smaller than Thr. It can take in both pyruvate and reduced NADH, even when water enters the pocket under high hydrostatic pressure, whereas that of fish dwelling in shallow habitats cannot. The larger pocket structure would be vulnerable to high pressure. The amino acid sequences of hagfish LDHs were deduced from the nucleotide sequences obtained.
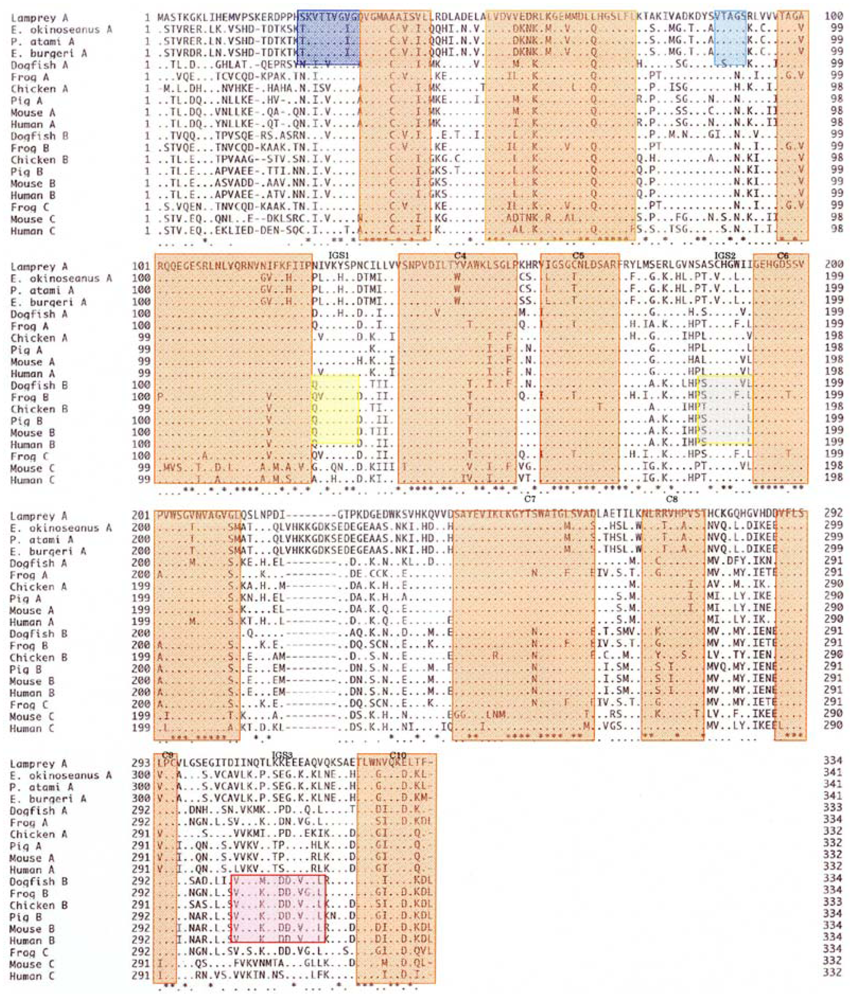
The amino acid sequences of 341 residues in the skeletal muscle LDHs in the three hagfishes were compared with those of previously reported LDHs. As seen in
Figure 6, the hagfish LDH-As had a specific insertion of ten amino acids (221–230). There are ten regions (C1–C10) common to LDHs from the vertebrates examined, and two Cyclostomata-specific regions (S1 and S2) have high homogeny (89–100%). Three regions (IGS1, IGS2, and IGS3) altered their structures during the differentiation of LDH isozymes, and those regions remain in vertebrate LDH-B with 67–86% homology.
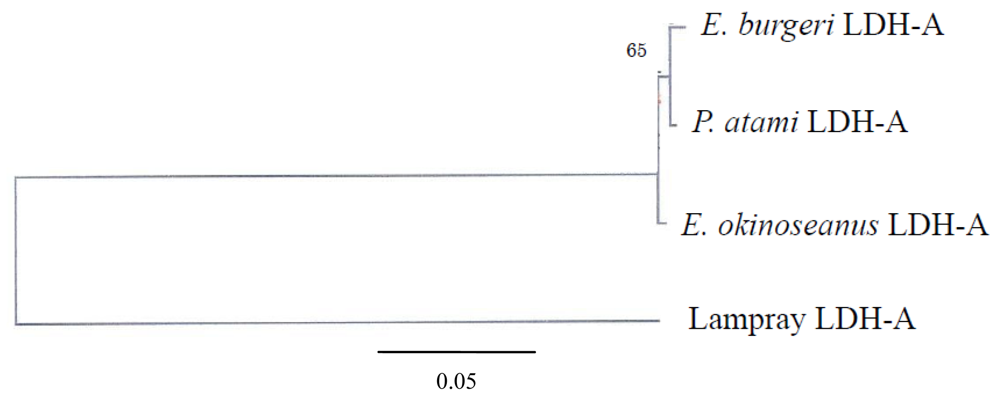
The phylogenetic tree of Cyclostomata LDH genes inferred from the present results suggests that LDH-A of hagfish diverged just after that of lampreys (
Figure 7). Although they probably evolved from a single ancestral gene, the genes encoding LDH isoenzymes are now quite distinct. The mechanism of evolution of LDH-A to LDH-B may be clarified through further investigation of the most prototypical LDH-B in hagfish.
During the evolution from Bacteria to Cyclostomata, residues 1–22 at the
N-terminus were added. IGS2 varies from a loop structure to a β-sheet structure. At the same time, IGS3 changed from a loop structure into a β-sheet, and it changed again to a loop structure during the evolution to dogfish LDH-A. The β-sheet structure remains in LDH-B. Fifty-seven percent of the amino acids of cyclostomata IGS3, 43% of dogfish LDH-A, 86% of dogfish LDH-B, and 14% of human LDH-A are in a β-sheet, while all (100%) of human LDH-B amino acids are postulated to be in a β-sheet. The three-dimensional structures presented in
Figure 8 suggest that Cyclostomata IGS3 has properties intermediate between those of A and B before it differentiates into the A and B types. This tendency was similar in IGS1 and IGS2.
4. Conclusions
The LDHs of the skeletal muscle from the three hagfishes E. burgeri, E. okinoseanus, and M. garmani, which are the lowest extant vertebrates, were purified and their properties were examined. The hagfish were revealed to have 10 specific additional amino acids, for a total of 341. The additional sequence (221–230) is located next to the hinge position (219) to introduce NAD and pyruvate. The additional sequence may cause less flexibility at the hinge position, resulting in the heat instability of hagfish LDHs. The optimal pH values (8.5–9.5) in the reaction from lactic acid to pyruvic acid of hagfish LDHs were almost the same as those of human (8.8) and cod (9.0) LDHs, although the optimum pH values (6.0–6.2) in the reaction from pyruvic acid to lactate acid of hagfishes were more acidic than those of human (7.2) and cod (7.5) LDHs. Differences in the three hagfish LDHs were apparent in the direction of the reversible reactions.
In the Cyclostomata, E. japonica has been shown to have a single LDH subunit, while hagfish have two subunits, LDH-A and LDH-B. Thus, hagfish LDH-B is the most primitive form examined. During the evolution from bacteria to lampreys, residues 1–22 at the N-terminus were added. This region is the part where the subunits connect with each other and changes from hydrophobic to hydrophilic during evolution. It is assumed that changes in this part caused the diversification of the isozymes.
The effects of high hydrostatic pressure on LDH activity were examined under pressures from 0.1 to 100 MPa. The activity of LDH-A4 from E. burgeri (shallow-sea species) was lost at pressure greater than 5 MPa. The activity of LDH-A4 from P. atami (inhabiting depths of ~400 m) was maintained at pressure of 10 MPa, but decreased to about 55% at 15 MPa and to about 15% at 20 MPa. Among the three hagfish, E. okinoseanus inhabits the deepest waters at around 800–1000 m. Its LDH-A4 appeared to be fully active at pressures up to 40 MPa and it maintained 70% of that activity even at 100 MPa. These results suggest that the LDH-A4 of E. okinoseanus is more adapted to high-pressure conditions than that of the other hagfishes.
Three possible mechanisms can be proposed as the reason for the inactivation of hagfish LDHs at high hydrostatic pressure: (1) the dissociation of the LDHs into four monomers; (2) changes in the structures near active sites; and (3) an increase in the activation volumes of LDHs. Six amino acid substitutions between the three LDH primary structures were found. Among them, two amino acid residues are likely to be responsible for the high-pressure resistance of
E. okinoseanus LDH. On the LDHs from both deep-sea and shallow living species,
Corphaenoides armatus and
Gadus morhua, the results of circular dichroism spectroscopic analysis were reported protein unfolding as the cause of inactivation at high pressure [
9]. The differences in the 6th amino acid residue of the hagfish LDHs would affect their pressure resistance. The results of electrophoresis under high-pressure conditions show that the dissociation of tetramers caused the inactivation of
E. burgeri LDH [
12]. To confirm that the predicted effects of substitutions indeed occur, we are performing site-directed mutagenesis. This method is the replacement of Ala6 with Glu in
E. okinoseanus LDH (now in progress).
Although the cDNAs of the LDH-A of the three hagfishes were clarified in the genetic study, hagfish LDH-B, which diverged from the lamprey LDH and achieved molecular variety for the first time, has not been elucidated. Thus, the amino acid sequences of those LDH-B and their enzymatic properties should be examined. The phylogenetic tree of Cyclostomata LDH-A genes was demonstrated in this study, but the relationship between Cyclostomata LDH and those of the Bacteria is not yet completely understood. The LDH-As of both the Ascidians and the Amphioxus, which are located just before the Cyclostomata in the phylogenetic tree, would be key in the study of evolution. The compositions of LDH isozymes are reported to change in human diseases [
24]. The relationship between the pressure tolerance of LDH and its isozyme composition is expected to contribute to the diagnosis of some human diseases.
Evolutionary medicine studies diseases from the viewpoint of evolution. We propose herein a new field, molecular evolutionary medicine, which focuses on genetic evolution [
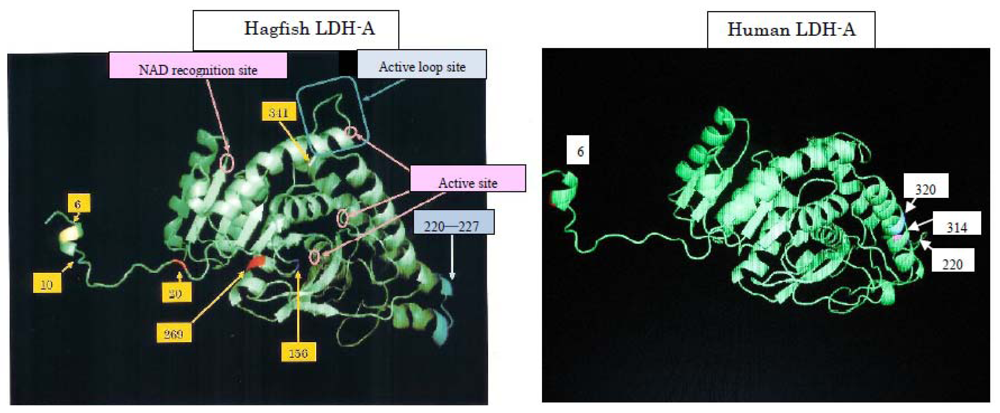
25]. We are comparing genetic variations in relation to human diseases. As an example, human and hagfish LDHs are related (
Figure 9). When subjected to physical pressure, those LDHs exhibit new properties. The amino acids responsible for pressure tolerance are the same in both human and hagfish LDHs, and substitutions of these amino acids occurred as adaptations during evolution. Mutations in these amino acids can result in anomalies that may be implicated in the development of human diseases. We are developing a device that can measure LDH activity in the blood of the patient with 6th amino acid mutation at around three MPa, which is below the pressure causing the dissociation of rabbit LDH-As and pig LDH-Bs (data not shown).