Quality not Quantity: The Role of Marine Natural Products in Drug Discovery and Reverse Chemical Proteomics
Abstract
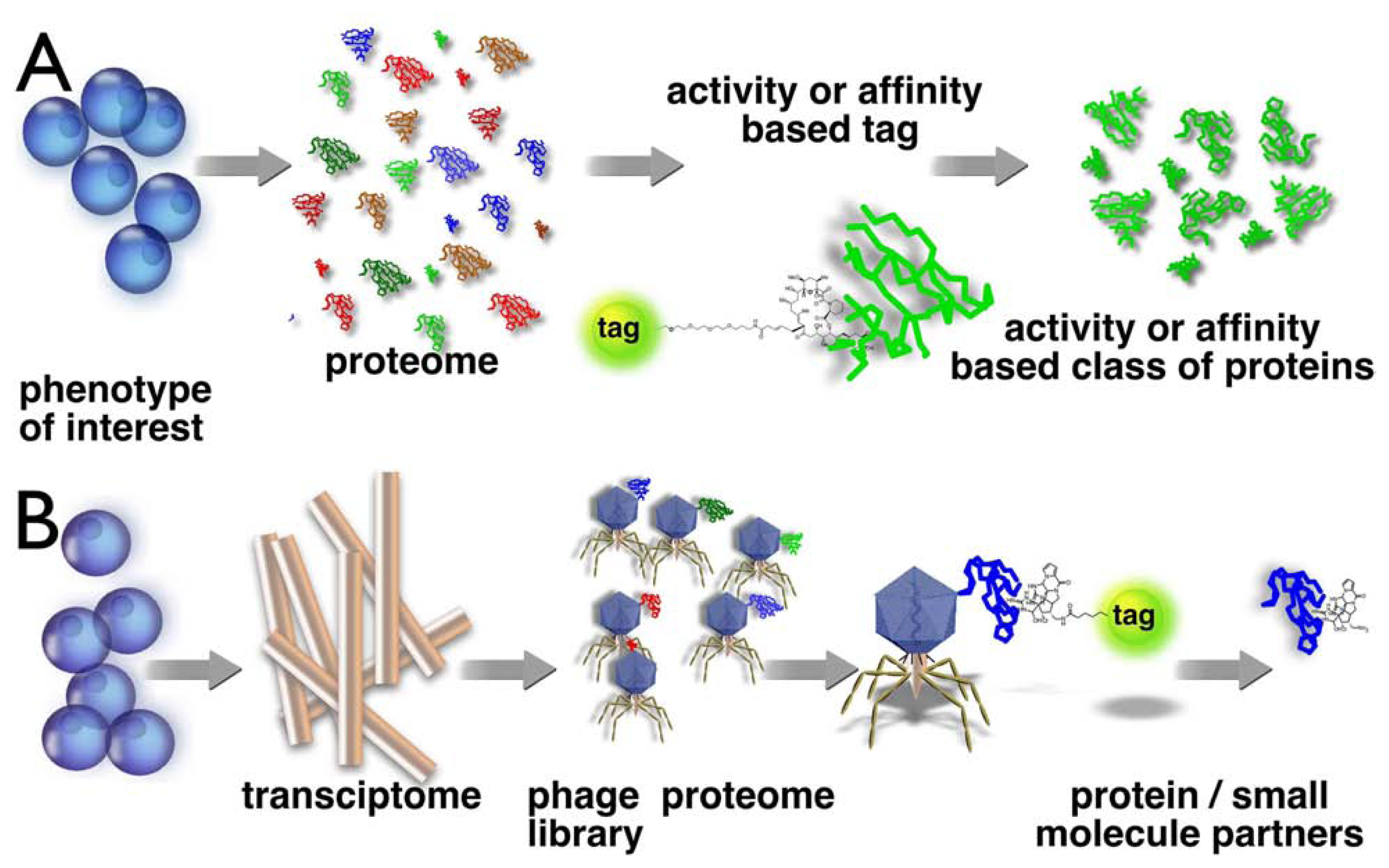
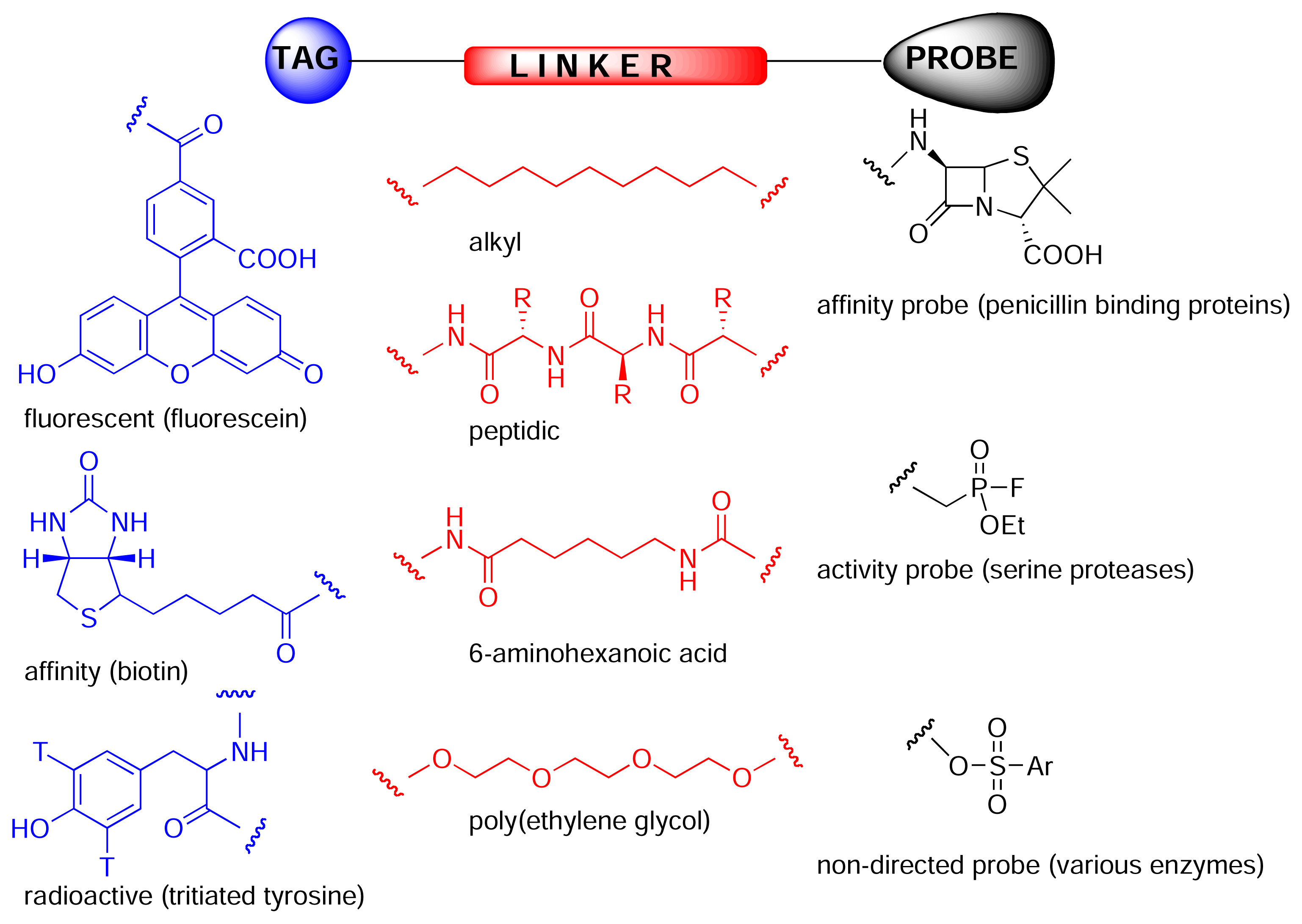
:Introduction
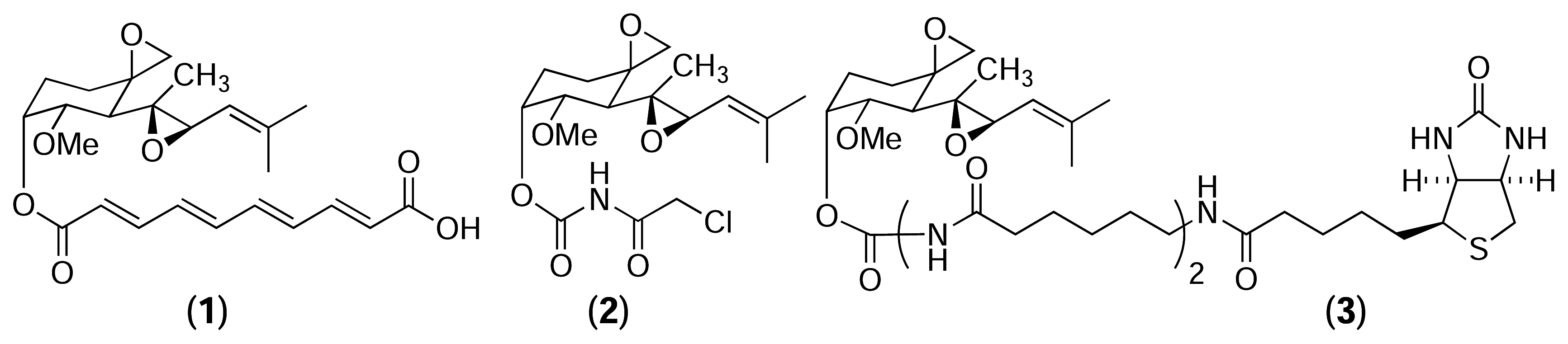

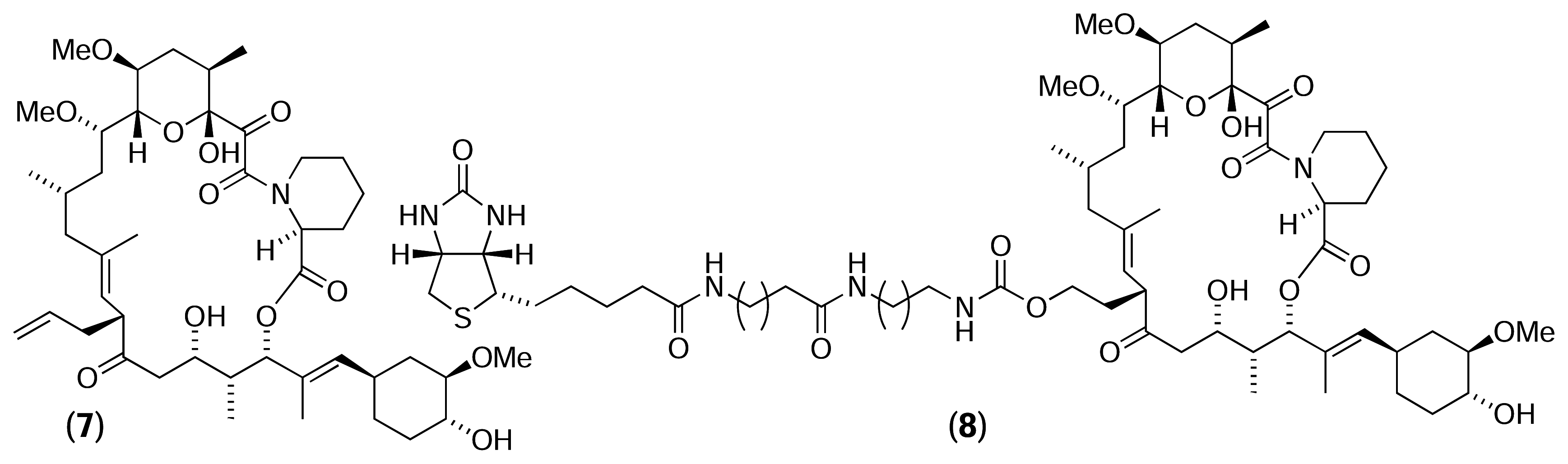
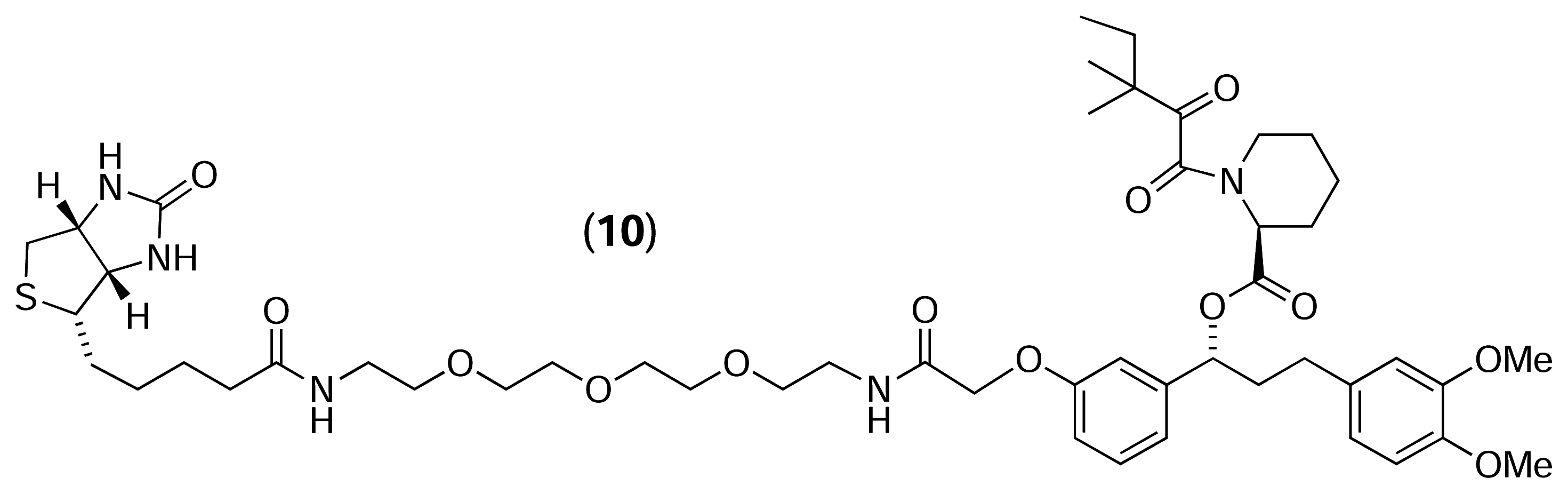
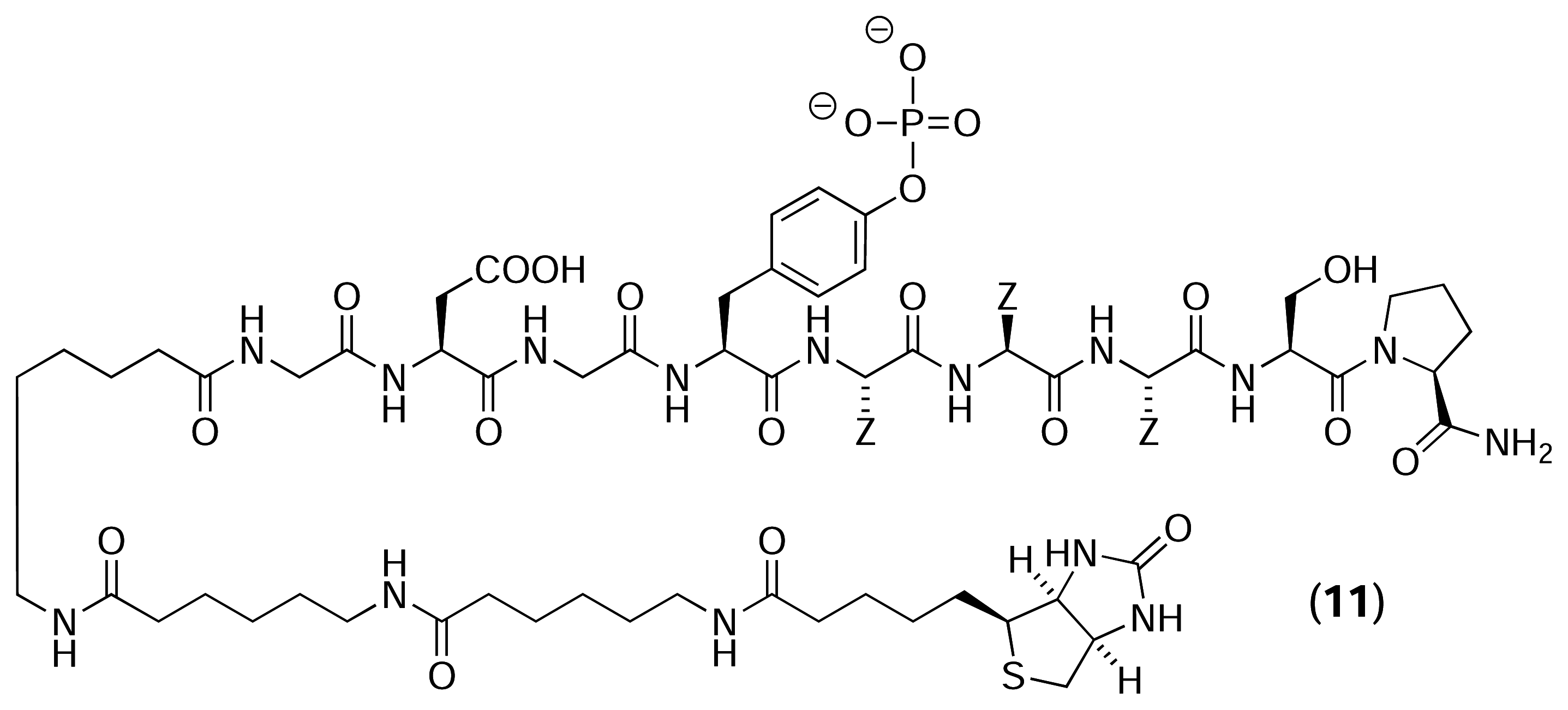

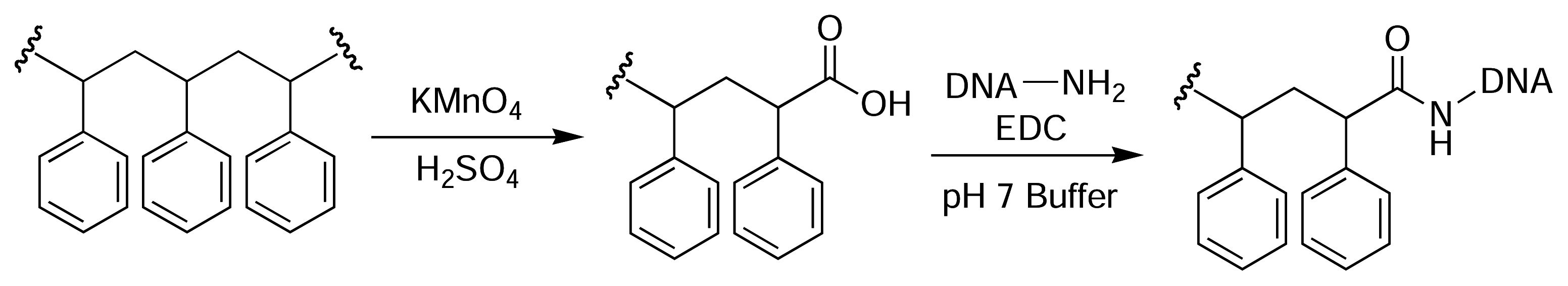
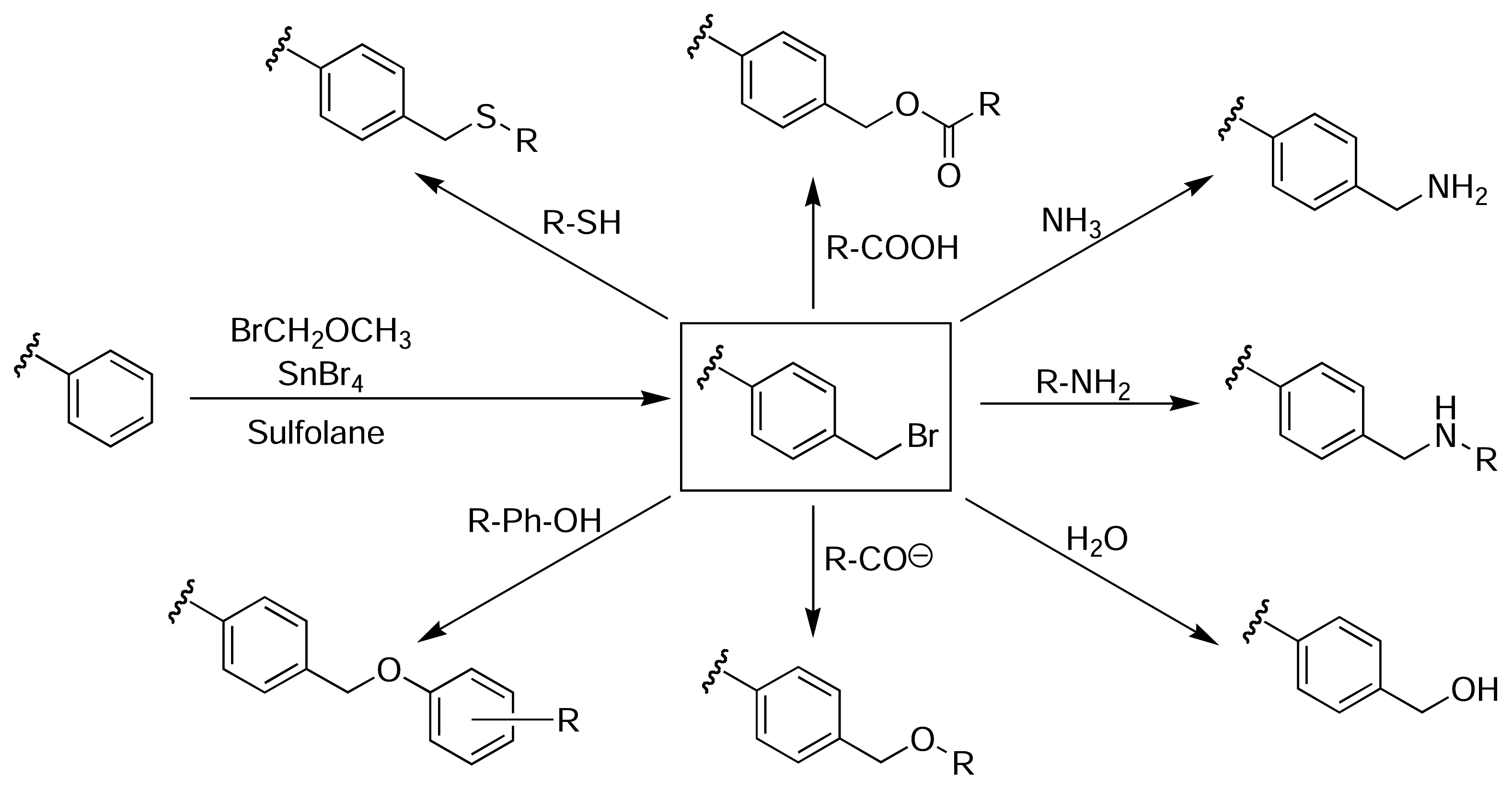
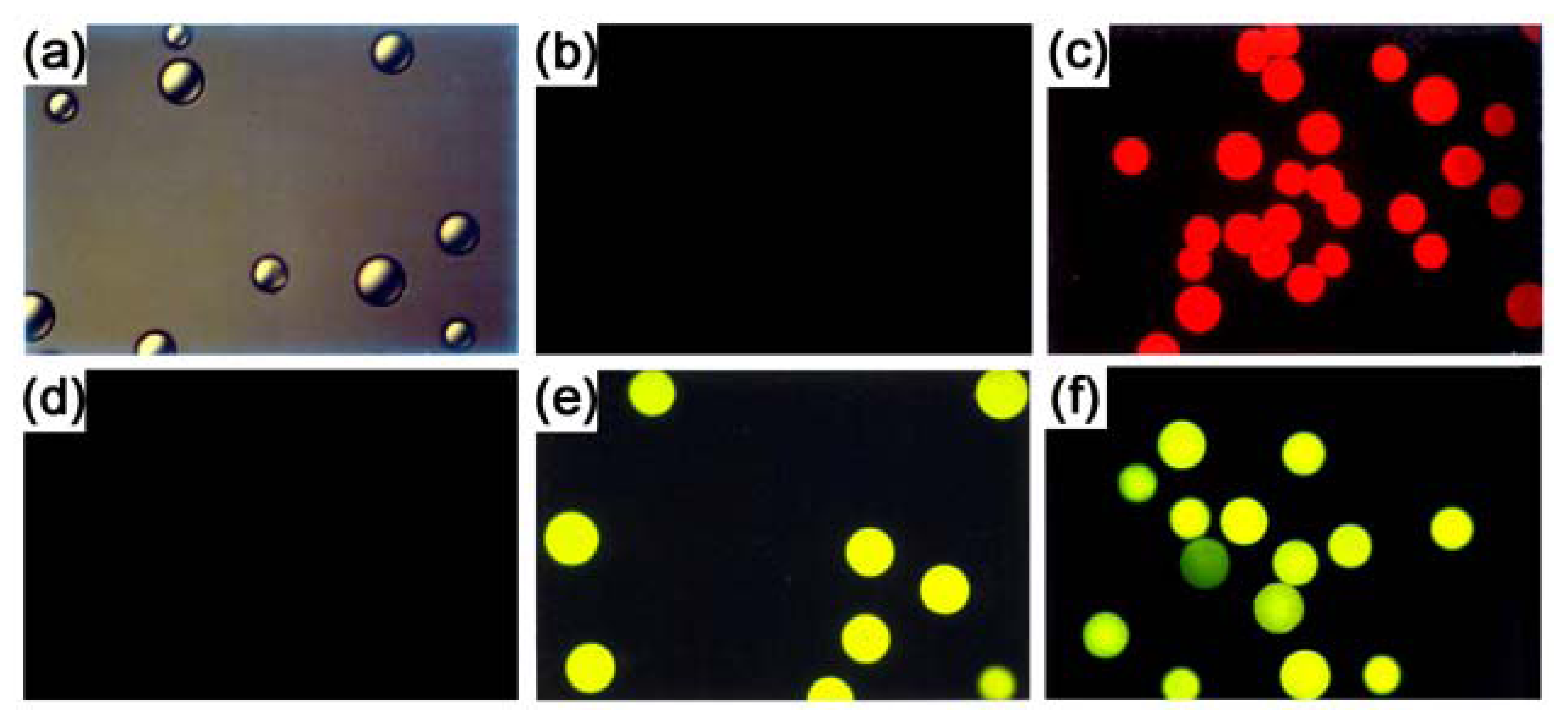
Results and Discussion
Experimental
General
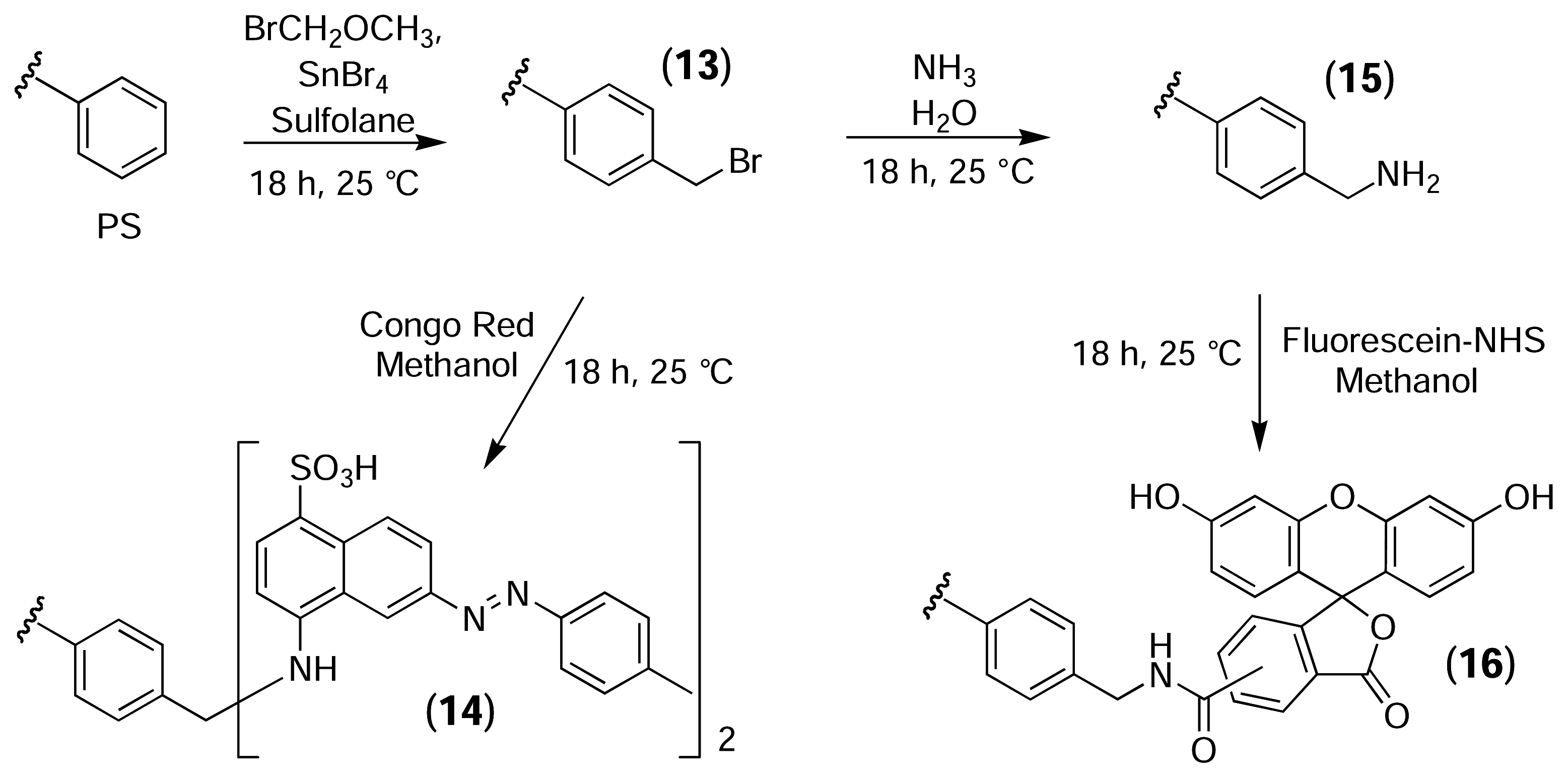
Derivatisation of Polystyrene (PS) Resins
PS-Bromomethyled Resin (13)
PS-Congo Red Resin (14)
PS-NH2 Resin (15)
PS-Fluorescein Resin (16)
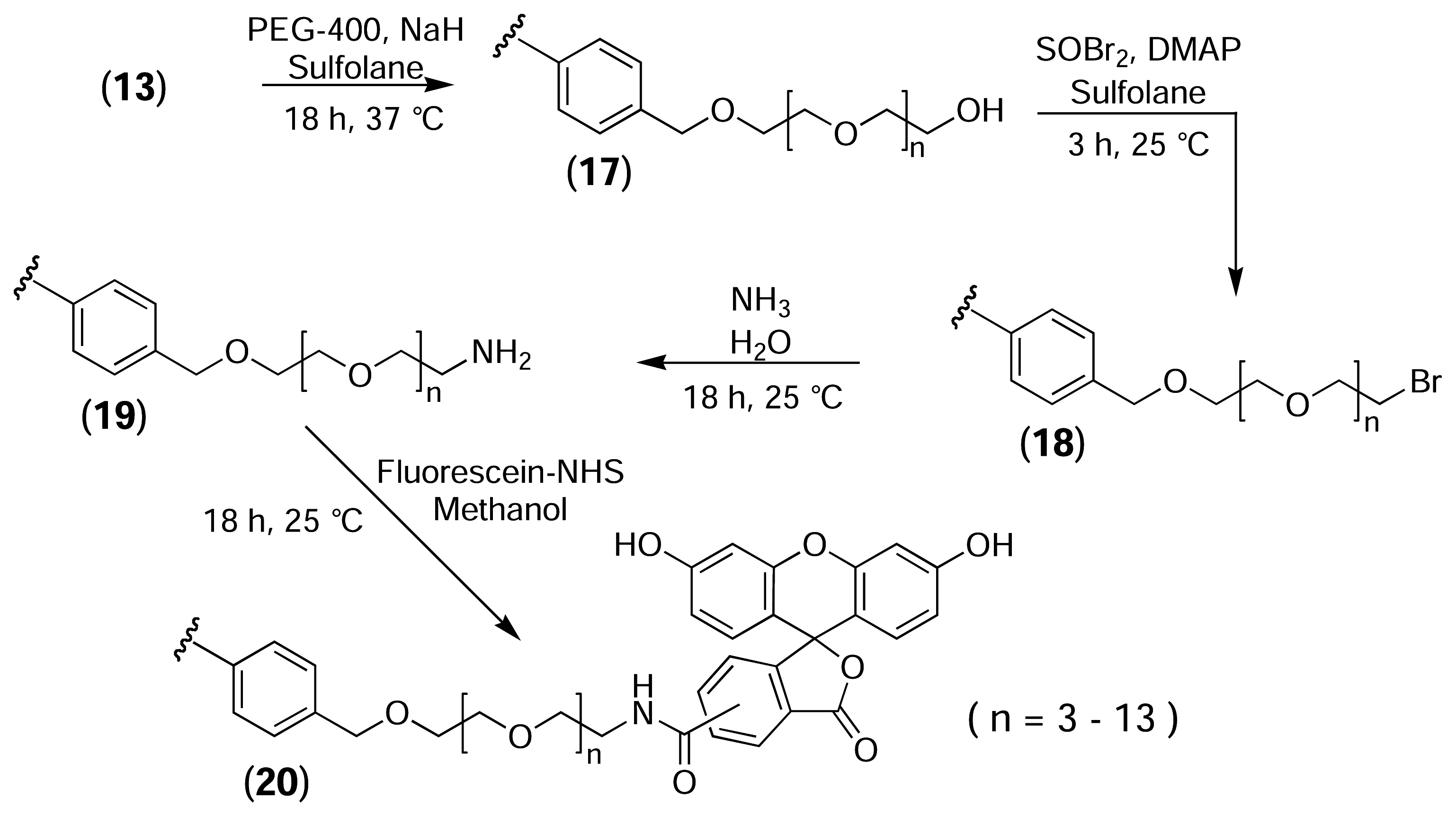
PS-PEG-OH Resin (17)
PS-PEG-Br Resin (18)
PS-PEG-NH2 Resin (19)
PS-Bromomethylated Plate (13)
PS-PEG-OH Plate (17)
PS-PEG-Br Plate (18)
PS-NH2 Plate (15)
PS-PEG-NH2 Plate (19)
PS-PEG-S-S-OSu Plate (21)
PS-PEG-S-S-Palau’amine Plate (22)
Phage Display
Preparation of Bacterial Cultures
Growth of T7 Lysates
Affinity Selections
Picking Plaques
Amplification, Sequencing and Fingerprinting of cDNA Inserts
Gel Electrophoresis
DNA Sequencing




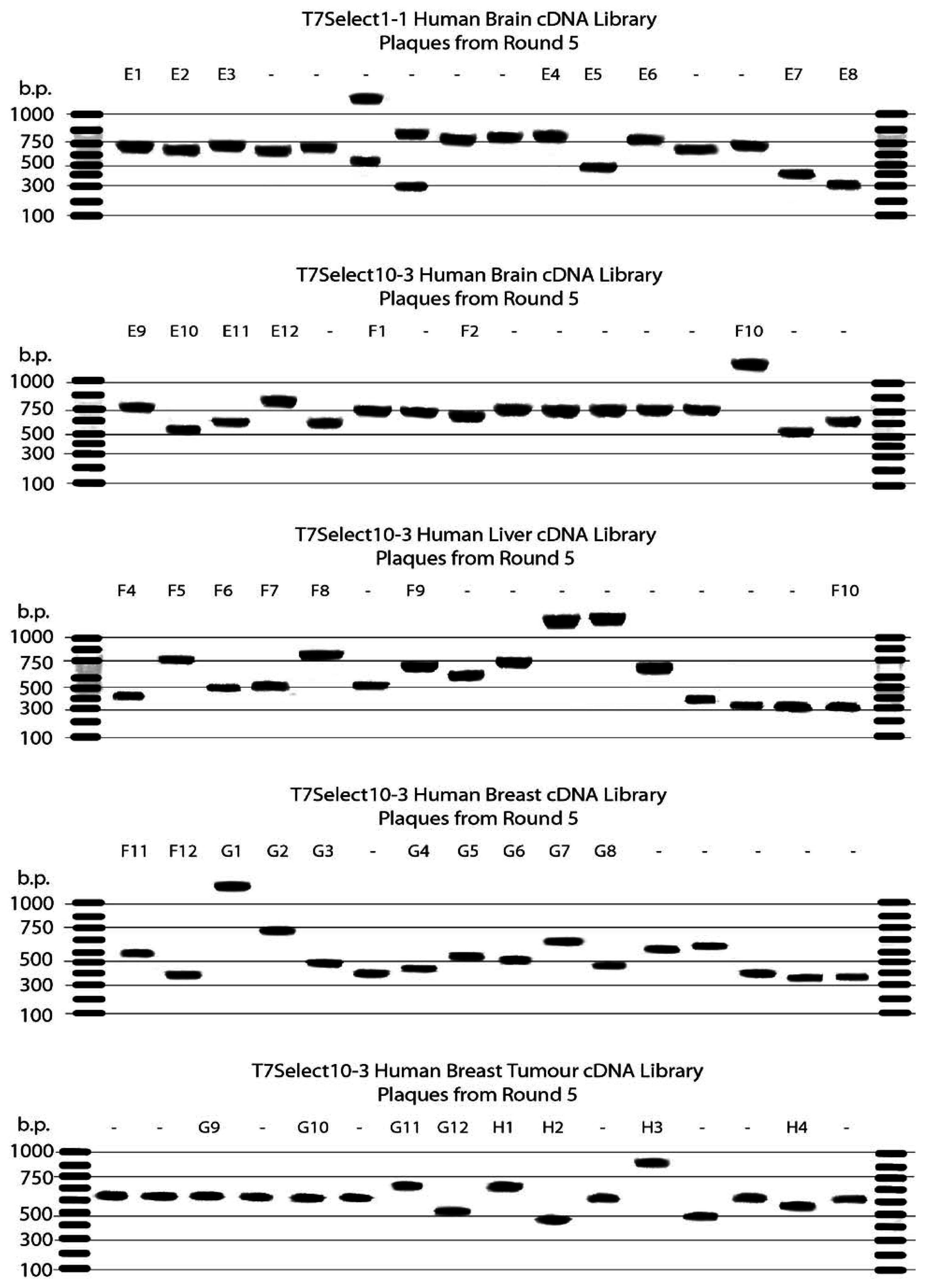
| Library | Plaques | Gene Identified from DNA Sequence | Notes |
|---|---|---|---|
| 1-1 Brain | E1, E3, E6 | Complete mitochondrial genome, haplotype B4a2, individual Ya015 | Fragment In Backwards |
| 1-1 Brain | E2 | Prader-Willi/Angelman syndrome region on chromosome 15 | Fragment |
| 1-1 Brain | E4 | Homo sapiens Chromosome 16 BAC clone CIT987SK-44M2 | Fragment |
| 1-1 Brain | E5 | Human DNA sequence from clone RP11-364G4 on chromosome 13 | Fragment |
| 1-1 Brain | E7 | Homo sapiens cDNA FLJ42917 fis, clone BRHIP3026335. | Fragment |
| 1-1 Brain | E8 | Human DNA sequence from clone RP11-4F1 on chromosome 10 | Fragment In Backwards |
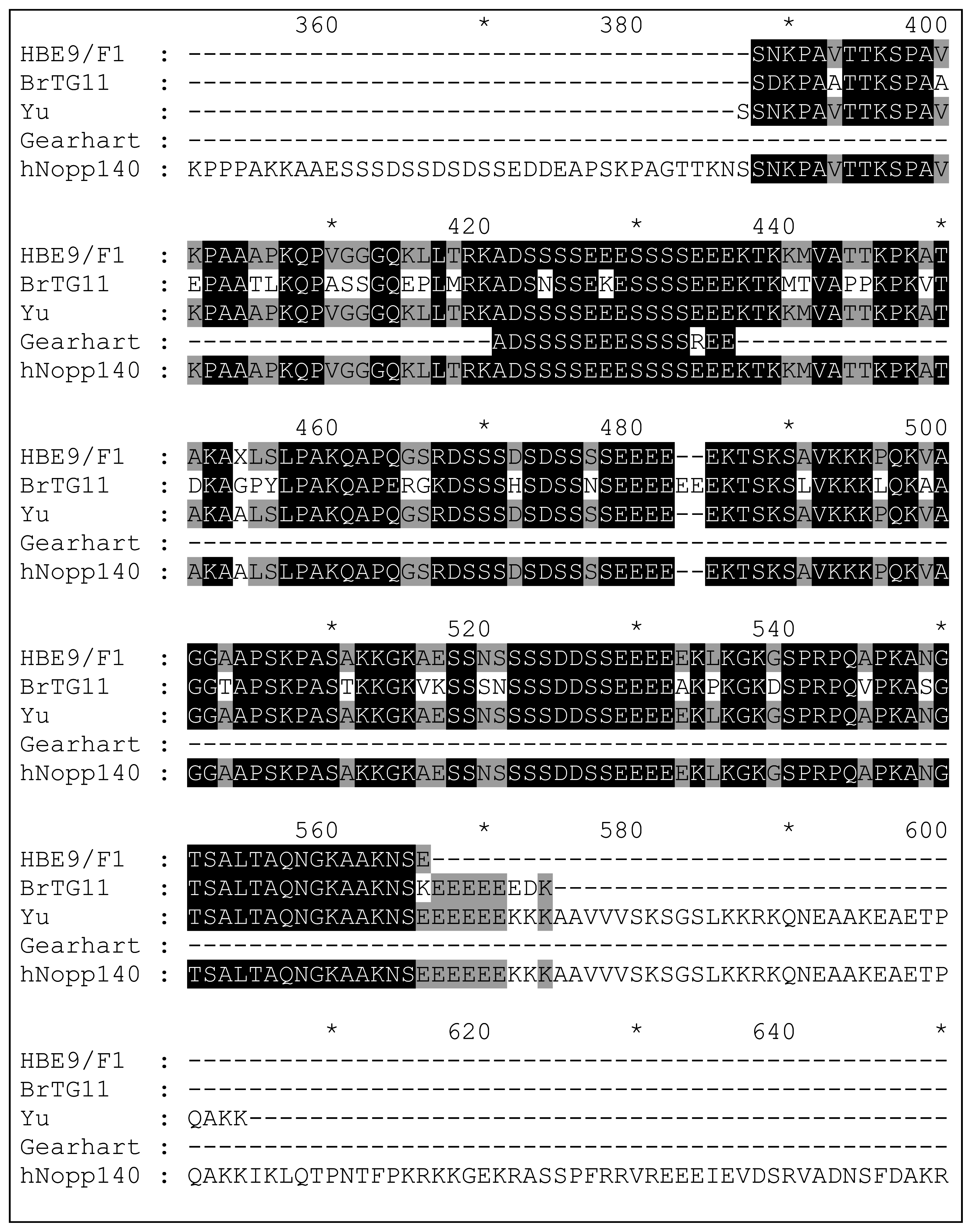
| 10-3 Brain | E9, F1 | Homo sapiens nucleolar and coiled-body phosphoprotein 1 (hNopp140) | Fragment In Frame |
| 10-3 Brain | E10 | Homo sapiens BAC clone RP11-696N14 from 4 | Fragment In Backwards |
| 10-3 Brain | E11 | Complete mitochondrial genome, haplotype B4a1a individual Am145. | Fragment In Backwards |
| 10-3 Brain | E12 | Leucine zipper, putative tumour suppressor 1 (LZTS1) | Fragment Not in CDS |
| 10-3 Brain | F2 | Homo sapiens phosphatase and actin regulator 3 (PHACTR3) | Fragment In Backwards |
| Liver | F3 | ATP synthase, H+transporting | Contamination |
| Liver | F4 | Diazepam binding inhibitor (GABA receptor modulator) | Fragment Not in CDS |
| Liver | F5 | Homo sapiens chromosome clone RP11-323I15 | Fragment In Backwards |
| Liver | F6 | Homo sapiens complement component 1, s subcomponent | Fragment Not in CDS |
| Liver | F7 | Human serum albumin (ALB) gene | Fragment |
| Liver | F8 | Homo sapiens 12 BAC RP11-218M22 | Fragment |
| Liver | F9 | Homo sapiens cDNA FLJ11946 fis, clone HEMBB1000709 | Fragment In Backwards |
| Liver | F10 | Homo sapiens ferritin light polypeptide | Fragment Frame 3 |
| Breast | F11 | Homo sapiens splicing factor 3b, subunit 2 (SF3B2) | Fragment Frame 2 |
| Breast | F12 | Homo sapiens chromosome 5 clone RP11-394O4 | Fragment |
| Breast | G1 | ATP synthase, H+transporting | Contamination |
| Breast | G2 | Homo sapiens snRNA activating protein complex (SNAP190) | Fragment Frame 3 |
| Breast | G3 | Homo sapiens ribosomal protein L10 (RPL10) | Fragment Frame 2 |
| Breast | G4 | Homo sapiens metastasis associated lung adenocarcinoma | Fragment |
| Breast | G5 | Lectin, galactoside-binding, soluble, 1 (galectin 1) | Fragment Frame 3 |
| Breast | G6 | Homo sapiens isolate H5-08 mitochondrion | Fragment |
| Breast | G7 | Human DNA sequence from clone RP11-337N19 on chromosome 10 | Fragment |
| Breast | G8 | Human mRNA for erythrocyte adducin alpha subunit | Fragment Not in CDS |
| Breast Tumour | G9, G10 | Heterogeneous nuclear ribonucleoprotein H3 (2H9)(HNRPH3) | Fragment Frame 3 |
| Breast Tumour | G11 | Homo sapiens nucleolar phosphoprotein p130 (hNopp140) | Fragment In Frame |
| Breast Tumour | G12 | Homo sapiens SMT3 suppressor of mif two 3 homolog 2 | Fragment Frame 2 |
| Breast Tumour | H1 | Homo sapiens BAC clone RP11-572N21 from 2 | Fragment |
| Breast Tumour | H2 | Homo sapiens cDNA FLJ26671 fis, clone MPG03325. | Fragment In Backwards |
| Breast Tumour | H3 | Chromosome 14 clone RP11-45G3 containing neurexin III gene | Fragment |
| Breast Tumour | H4 | Human DNA sequence from clone RP11-84A14 on chromosome 1 | Fragment |
| Charged | Polar | Intermediate | Hydrophobic | ||||
|---|---|---|---|---|---|---|---|
| Arg | 5.0% | Asn | 3.0% | Ala | 12.0% | Ile | 1.3% |
| Asp | 3.7% | Gln | 4.0% | Cys | 0.0% | Leu | 3.0% |
| Glu | 11.4% | Ser | 15.4% | Gly | 6.7% | Phe | 2.0% |
| His | 0.3% | Thr | 4.3% | Met | 0.3% | Trp | 0.3% |
| Lys | 17.1% | Pro | 5.4% | Tyr | 0.3% | ||
| Val | 4.3% | ||||||
| TOTAL | 37.5% | TOTAL | 26.7% | TOTAL | 24.4% | TOTAL | 11.2% |
| CYCLES | TEMPERATURE | TIME |
|---|---|---|
| 1 | 94 °C | 2 min 30 s |
| 94 °C | 45 s | |
| 35 | 55 °C | 60 s |
| 72 °C | 30 s | |
| 1 | 72 °C | 10 min |
| 1 | 4 °C | ∞ |
Acknowledgements
- Sample availability: Not available.
Reference and Notes
- Rouhi, A. M. Rediscovering Natural Products. Chem. Eng. News 2003, 81, 77–91. [Google Scholar]
- Newman, D. J.; Cragg, G. M.; Snader, K. M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod 2003, 66, 1022–1037. [Google Scholar]
- Feher, M.; Schmidt, J. M. Property distributions: Differences between drugs, Natural Products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci 2003, 43, 218–227. [Google Scholar]
- Adam, G. C.; Sorensen, E. J.; Cravatt, B. F. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat. Biotechnol 2002, 20, 805–809. [Google Scholar]
- Jeffery, D. A.; Bogyo, M. Chemical proteomics and its application to drug discovery. Curr. Opin. Biotechnol 2003, 14, 87–95. [Google Scholar]
- Patricelli, M. P.; Giang, D. K.; Stamp, L. M.; Burbaum, J. J. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 2001, 1, 1067–1071. [Google Scholar]
- Speers, A. E.; Cravatt, B. F. Chemical strategies for activity-based proteomics. ChemBioChem 2004, 5, 41–47. [Google Scholar]
- Zhao, G.; Meier, T. I.; Kahl, S. D.; Gee, K. R.; Blaszczak, L. C. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother 1999, 43, 1124–1128. [Google Scholar]
- Taylor, E. W. The mechanism of colchicine inhibition of mitosis. I. Kinetics of inhibition and the binding of colchicine-3H. J. Cell Biol 1965, 25, 145–160. [Google Scholar]
- Borisy, G. G.; Taylor, E. W. The mechanism of action of colchicine. Binding of colchicine-3H to cellular protein. J. Cell Biol 1967, 34, 525–533. [Google Scholar]
- Borisy, G. G.; Taylor, E. W. The mechanism of action of colchicine. Colchicine binding to sea urchin eggs and the mitotic apparatus. J. Cell Biol 1967, 34, 535–548. [Google Scholar]
- Shelanski, M. L.; Taylor, E. W. Isolation of a protein subunit from microtubules. J. Cell Biol 1967, 34, 549–554. [Google Scholar]
- Weisenberg, R. C.; Broisy, G. G.; Taylor, E. W. Colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry 1968, 7, 4466–4479. [Google Scholar]
- Piggott, A. M.; Karuso, P. Quality, not quantity: The role of natural products and chemical proteomics in modern drug discovery. Comb. Chem. High Throughput Screen 2004, 7, 607–630. [Google Scholar]
- Sin, N.; Meng, L.; Wang, M. Q. W.; Wen, J. J.; Bornmann, W. G.; Crews, C. M. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Nat. Acad. Sci. USA 1997, 94, 6099–6103. [Google Scholar]
- Sin, N.; Meng, L.; Auth, H.; Crews, C. M. Eponemycin analogues: syntheses and use as probes of angiogenesis. Bioorg. Med. Chem 1998, 6, 1209–1217. [Google Scholar]
- Abe, J.; Zhou, W.; Takuwa, N.; Taguchi, J.; Kurokawa, K.; Kumada, M.; Takuwa, Y. A fumagillin derivative angiogenesis inhibitor, AGM-1470, inhibits activation of cyclin-dependent kinases and phosphorylation of retinoblastoma gene product but not protein tyrosyl phosphorylation or protooncogene expression in vascular endothelial cells. Cancer Res 1994, 54, 3407–3412. [Google Scholar]
- Lowther, W. T.; McMillen, D. A.; Orville, A. M.; Matthews, B. W. The anti-angiogenic agent fumagillin covalently modifies a conserved active-site histidine in the Escherichia coli methionine aminopeptidase. Proc. Nat. Acad. Sci. USA 1998, 95, 12153–12157. [Google Scholar]
- Liu, S.; Widom, J.; Kemp, C. W.; Crews, C. M.; Clardy, J. Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science 1998, 282, 1324–1327. [Google Scholar]
- Luibrand, R. T.; Erdman, T. R.; Vollmer, J. J.; Scheuer, P. J.; Finer, J.; Clardy, J. Ilimaquinone, a sesquiterpenoid quinone from a marine sponge. Tetrahedron 1979, 35, 609–612. [Google Scholar]
- Takizawa, P. A.; Yucel, J. K.; Veit, B.; Faulkner, D. J.; Deerinck, T.; Soto, G.; Ellisman, M.; Malhotra, V. Complete vesiculation of Golgi membranes and inhibition of protein transport by a novel sea sponge metabolite, ilimaquinone. Cell 1993, 73, 1079–1090. [Google Scholar]
- Veit, B.; Yucel, J. K.; Malhotra, V. Microtubule independent vesiculation of Golgi membranes and the reassembly of vesicles into Golgi stacks. J. Cell Biol 1993, 122, 1197–1206. [Google Scholar]
- Radeke, H. S.; Snapper, M. L. Photoaffinity study of the cellular interactions of ilimaquinone. Bioorg. Med. Chem 1998, 6, 1227–1232. [Google Scholar]
- Radeke, H. S.; Digits, C. A.; Casaubon, R. L.; Snapper, M. L. Interactions of (−)-ilimaquinone with methylation enzymes: implications for vesicular-mediated secretion. Chem. Biol 1999, 6, 639–647. [Google Scholar]
- Lee, S. Y.; Choi, J. H.; Xu, Z. Microbial cell-surface display. Trends Biotech 2003, 21, 45–52. [Google Scholar]
- Kondo, A.; Ueda, M. Yeast cell-surface display - applications of molecular display. Appl. Microbiol. Biotechnol 2004, 64, 28–40. [Google Scholar]
- Crews, C. M.; Collins, J. L.; Lane, W. S.; Snapper, M. L.; Schreiber, S. L. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1α. J. Biol. Chem 1994, 269, 15411–15414. [Google Scholar]
- Sche, P. P.; McKenzie, K. M.; White, J. D.; Austin, D. J. Display cloning: functional identification of natural product receptors using cDNA-phage display. Chem. Biol 1999, 6, 707–716. [Google Scholar]
- Sche, P. P.; McKenzie, K. M.; White, J. D.; Austin, D. J. Display cloning: Functional identification of natural product receptors using cDNA-phage display. [Erratum to document cited in CA132:45554]. Chem. Biol 2001, 8, 399–400. [Google Scholar]
- McKenzie, K. M.; Videlock, E. J.; Splittgerber, U.; Austin, D. J. Simultaneous identification of multiple protein targets by using complementary-DNA phage display and a Natural-Product-mimetic probe. Angew. Chem. Int. Ed 2004, 43, 4052–4055. [Google Scholar]
- Siekierka, J. J.; Hung, S. H. Y.; Poe, M.; Lin, C. S.; Sigal, N. H. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 1989, 341, 755–757. [Google Scholar]
- Harding, M. W.; Galat, A.; Uehling, D. E.; Schreiber, S. L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 1989, 341, 758–760. [Google Scholar]
- Videlock, E. J.; Chung, V. K.; Mohan, M. A.; Strok, T. M.; Austin, D. J. Two-dimensional diversity: Screening human cDNA phage display libraries with a random diversity probe for the display cloning of phosphotyrosine binding domains. J. Am. Chem. Soc 2004, 126, 3730–3731. [Google Scholar]
- Videlock, E. J.; Chung, V. K.; Hall, J. M.; Hines, J.; Agapakis, C. M.; Austin, D. J. Identification of a molecular recognition role for the activation loop phosphotyrosine of the Src tyrosine kinase. J. Am. Chem. Soc. 2005, 127. [Google Scholar]
- Shim, J. S.; Lee, J.; Park, H-J.; Park, S-J.; Kwon, H. J. A new curcumin derivative, HBC, interferes with the cell cycle progression of colon Cancer cells via antagonization of the Ca2+/calmodulin function. Chem. Biol. 2004, 11, 1455–1463. [Google Scholar]
- Merrifield, R. B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc 1963, 85, 2149–2154. [Google Scholar]
- Delgado, M.; Janda, K. D. Polymeric supports for solid phase organic synthesis. Curr. Org. Chem 2002, 6, 1031–1043. [Google Scholar]
- Blaney, P.; Grigg, R.; Sridharan, V. Traceless solid-phase organic synthesis. Chem. Rev 2002, 102, 2607–2624. [Google Scholar]
- Guillier, F.; Orain, D.; Bradley, M. Linkers and cleavage strategies in solid phase organic synthesis and combinatorial chemistry. Chem. Rev 2000, 100, 2091–2157. [Google Scholar]
- James, I. W. Linkers for solid phase organic synthesis. Tetrahedron 1999, 55, 4855–4946. [Google Scholar]
- Bochet, C. G. Photolabile protecting groups and linkers. J. Chem. Soc. Perkin Trans. 1 2002, 2, 125–142. [Google Scholar]
- Plunkett, M. J.; Ellman, J. A. A Silicon-Based Linker for Traceless Solid-Phase Synthesis. J. Org. Chem 1995, 60, 6006–6007. [Google Scholar]
- Patek, M.; Lebl, M. Safety-catch and multiply cleavable linkers in solid-phase synthesis. Biopolymers 1998, 47, 353–363. [Google Scholar]
- Rotmans, J. P.; Delwel, H. R. Cross-linking of Schistosoma mansoni antigens and their covalent binding on the surface of polystyrene microtitration trays for use in the ELISA. J. Immunol. Methods 1983, 57, 87–98. [Google Scholar]
- Aleixo, J. A. G.; Swaminathan, B.; Minnich, S. A.; Wallshein, V. A. Enzyme immunoassay: binding of Salmonella antigens to activated microtiter plates. J. Immunoass 1985, 6, 391–407. [Google Scholar]
- Zammatteo, N.; Girardeaux, C.; Delforge, D.; Pireaux, J-J.; Remacle, J. Amination of polystyrene microwells: application to the covalent grafting of DNA probes for hybridization assays. Anal. Biochem. 1996, 236, 85–94. [Google Scholar]
- Clark, B. R. Derivatized polystyrene and other polymer supports for spectroscopic studies; Aventis Pharmaceuticals Holdings Inc.: USA, 2003; p. 7. [Google Scholar]
- Bora, U.; Chugh, L.; Nahar, P. Covalent immobilization of proteins onto photoactivated polystyrene microtiter plates for enzyme-linked immunosorbent assay procedures. J. Immunol. Methods 2002, 268, 171–177. [Google Scholar]
- Nahar, P.; Wali, N. M.; Gandhi, R. P. Light-induced activation of an inert surface for covalent immobilization of a protein ligand. Anal. Biochem 2001, 294, 148–153. [Google Scholar]
- Fleet, G. W. J.; Porter, R. R.; Knowles, J. R. Affinity labeling of antibodies with aryl nitrene as reactive group. Nature 1969, 224, 511–512. [Google Scholar]
- Ito, Y.; Chen, G.; Imanishi, Y. Photoimmobilization of insulin onto polystyrene dishes for protein-free cell culture. Biotechnol. Prog 1996, 12, 700–702. [Google Scholar]
- Eckert, A. W.; Gröbe, D.; Rothe, U. Surface-modification of polystyrene-microtitre plates via grafting of glycidylmethacrylate and coating of poly-glycidylmethacrylate. Biomaterials 2000, 21, 441–447. [Google Scholar]
- Larsson, P. H.; Eklund, A.; Johansson, S. G. O.; Larsson, K. Covalent binding of proteins to grafted plastic surfaces suitable for immunoassays. II. Picograms of IgE detected in BAL fluid in sarcoidosis. J. Immunol. Methods 1997, 210, 41–49. [Google Scholar]
- Larsson, P. H.; Johansson, S. G. O.; Hult, A.; Göthe, S. Covalent binding of proteins to grafted plastic surfaces suitable for immunoassays. J. Immunol. Methods 1987, 98, 129–135. [Google Scholar]
- Butler, J. E. Solid supports in enzyme-linked immunosorbent assay and other solid-phase immunoassays. Methods Mol. Med 2004, 94, 333–372. [Google Scholar]
- Kinnel, R. B.; Gehrken, H-P.; Scheuer, P. J. Palau’amine: a cytotoxic and immunosuppressive hexacyclic bisguanidine antibiotic from the sponge Stylotella agminata. J. Am. Chem. Soc. 1993, 115, 3376–3377. [Google Scholar]
- Kinnel, R. B.; Gehrken, H-P.; Swali, R.; Skoropowski, G.; Scheuer, P. J. Palau’amine and its congeners: A family of bioactive bisguanidines from the marine sponge Stylotella aurantium. J. Org. Chem. 1998, 63, 3281–3286. [Google Scholar]
- Garrido-Hernandez, H.; Nakadai, M.; Vimolratana, M.; Li, Q.; Doundoulakis, T.; Harran, P. G. Spirocycloisomerization of tethered alkylidene glycocyamidines: Synthesis of a base template common to the palau’amine family of alkaloids. Angew. Chem. Int. Ed 2005, 44, 765–769. [Google Scholar]
- Katz, J. D.; Overman, L. E. Studies towards the total synthesis of palau’amine. Formation of 4,5-dihydropyrrole-2-carboxylate intermediates by alkene-enamide ring-closing metathesis. Tetrahedron 2004, 60, 9559–9568. [Google Scholar]
- Dilley, A. S.; Romo, D. Enantioselective Strategy to the Spirocyclic Core of Palau’amine and Related Bisguanidine Marine Alkaloids. Org. Lett 2001, 3, 1535–1538. [Google Scholar]
- Overman, L. E.; Rogers, B. N.; Tellew, J. E.; Trenkle, W. C. Stereocontrolled synthesis of the tetracyclic core of the bisguanidine alkaloids Palau’amine and Styloguanidine. J. Am. Chem. Soc 1997, 119, 7159–7160. [Google Scholar]
- Meier, U. T.; Blobel, G. A nuclear localization signal binding protein in the nucleolus. J. Cell Biol 1990, 111, 2235–2245. [Google Scholar]
- Jin, Y.; Yu, J.; Yu, Y. G. Identifcation of hNopp140 as a binding partner for doxorubicin with phage display cloning method. Chem. Biol 2002, 9, 157–162. [Google Scholar]
- Gearhart, D. A. T.; Patricia, F.; Warren Beach, J. Identification of brain proteins that interact with 2-methylnorharman. An analog of the parkinsonian-inducing toxin MPP+. Neurosci. Res. 2002, 44, 255–265. [Google Scholar]
- Kim, Y-K.; Jin, Y.; Vukoti, K. M.; Park, J. K.; Kim, E. E.; Lee, K-J.; Yu, Y. G. Purification and characterization of human nucleolar phosphoprotein 140 expressed in Escherichia coli. Prot. Expr. Purif. 2003, 31, 260–264. [Google Scholar]
- Pai, C-Y.; Chen, H-K.; Sheu, H-L.; Yeh, N-H. Cell cycle-dependent alterations of a highly phosphorylated nucleolar protein p130 are associated with nucleologenesis. J. Cell Sci. 1995, 108, 1911–1920. [Google Scholar]
- Pai, C-Y.; Yeh, N-H. Cell proliferation-dependent expression of two isoforms of the nucleolar phosphoprotein p130. Biochem. Biophys. Res. Commun. 1996, 221, 581–587. [Google Scholar]
- Chen, H-K.; Yeh, N-H. The nucleolar phosphoprotein P130 is a GTPase/ATPase with intrinsic property to form large complexes triggered by F-and Mg2+. Biochem. Biophys. Res. Commun. 1997, 230, 370–375. [Google Scholar]
- Meier, U. T.; Blobel, G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 1992, 70, 127–138. [Google Scholar]
- Savinov, S. N.; Austin, D. J. The cloning of human genes using cDNA phage display and small-molecule chemical probes. Comb. Chem. High Throughput Screen 2001, 4, 593–597. [Google Scholar]
- Novagen, T7Select System Manual. USA. 2000, 1–21, Sample availability: Not available..
© 2005 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Piggott, A.M.; Karuso, P. Quality not Quantity: The Role of Marine Natural Products in Drug Discovery and Reverse Chemical Proteomics. Mar. Drugs 2005, 3, 36-63. https://doi.org/10.3390/md302036
Piggott AM, Karuso P. Quality not Quantity: The Role of Marine Natural Products in Drug Discovery and Reverse Chemical Proteomics. Marine Drugs. 2005; 3(2):36-63. https://doi.org/10.3390/md302036
Chicago/Turabian StylePiggott, Andrew M., and Peter Karuso. 2005. "Quality not Quantity: The Role of Marine Natural Products in Drug Discovery and Reverse Chemical Proteomics" Marine Drugs 3, no. 2: 36-63. https://doi.org/10.3390/md302036





