Polyketides and Alkaloids from the Marine-Derived Fungus Dichotomomyces cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus
Abstract
:1. Introduction
2. Results and Discussion
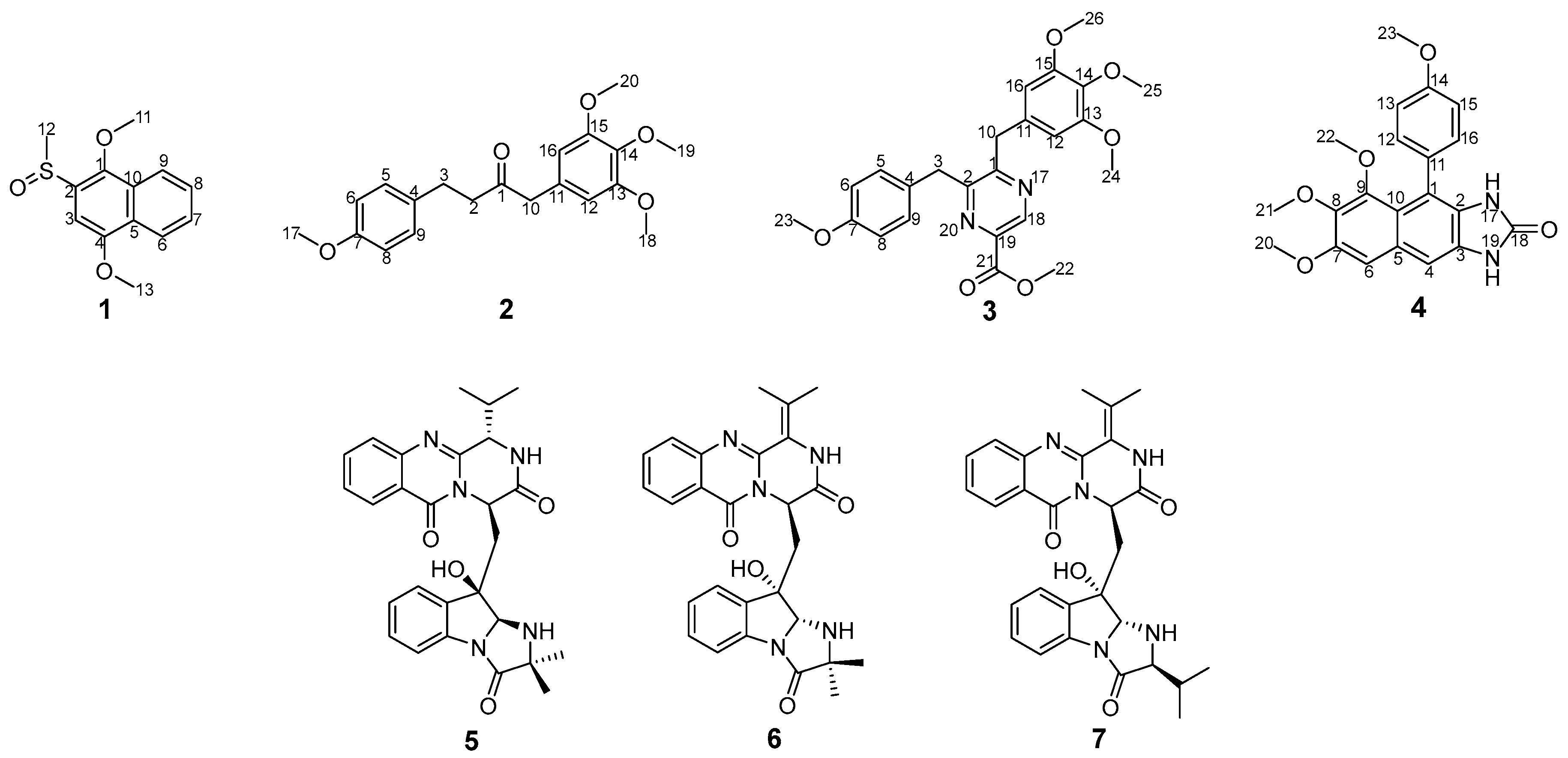
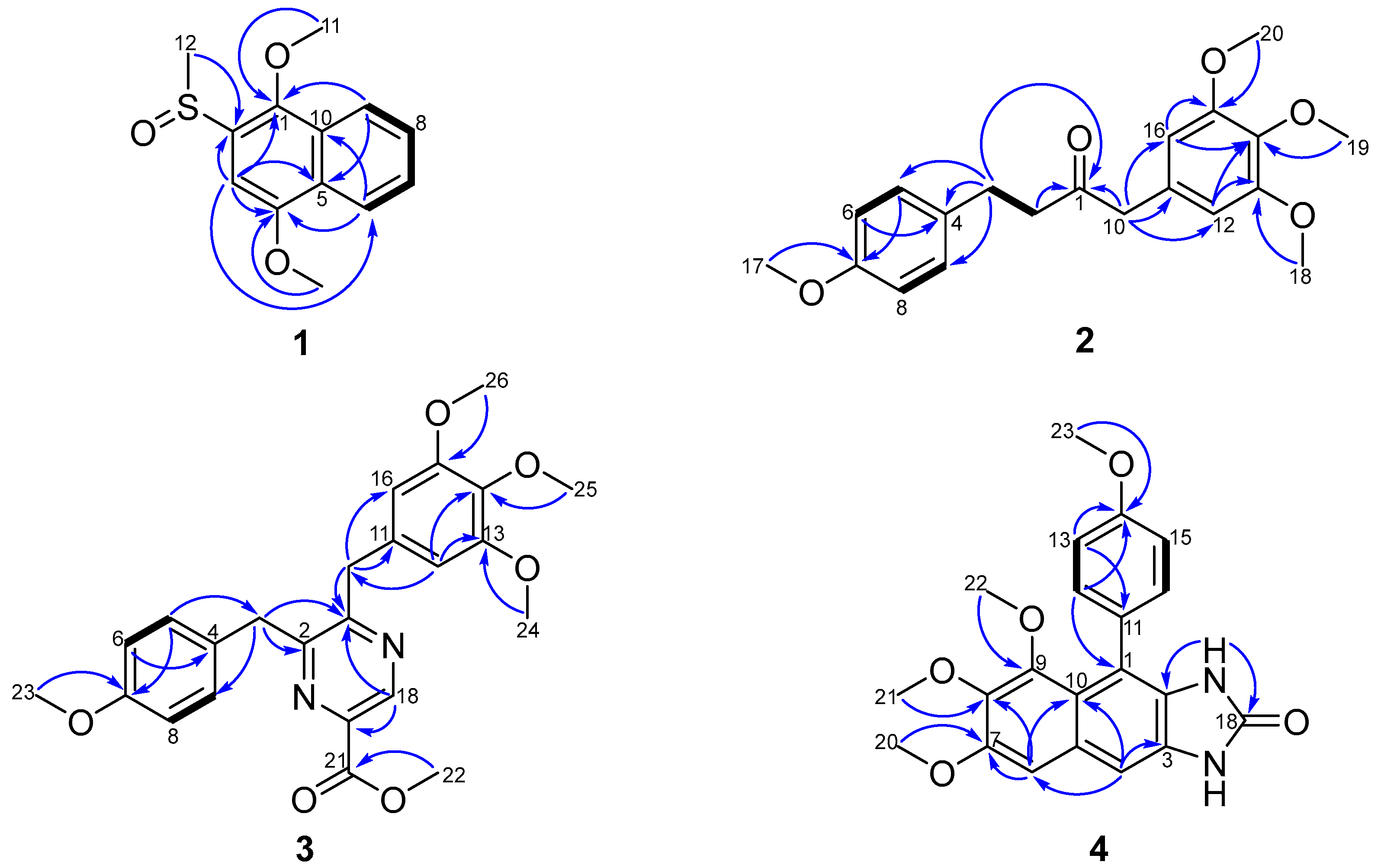
2.1. Structural Elucidation
2.2. Biological Activity
2.2.1. Cytotoxicity of Scequinadoline A (5), Quinadoline A (6), and Scequinadoline E (7)
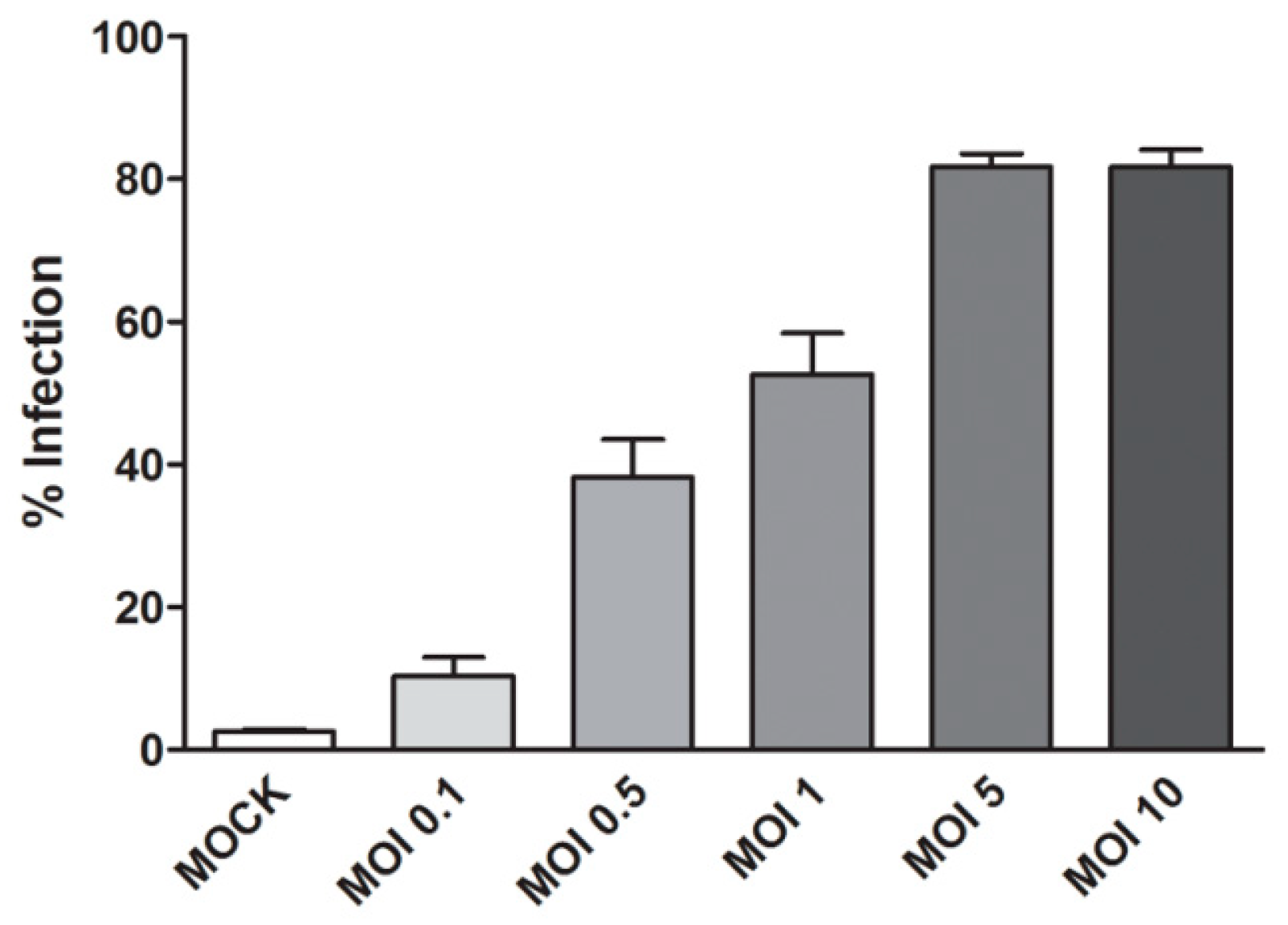
2.2.2. Optimization of DENV 2 Infection
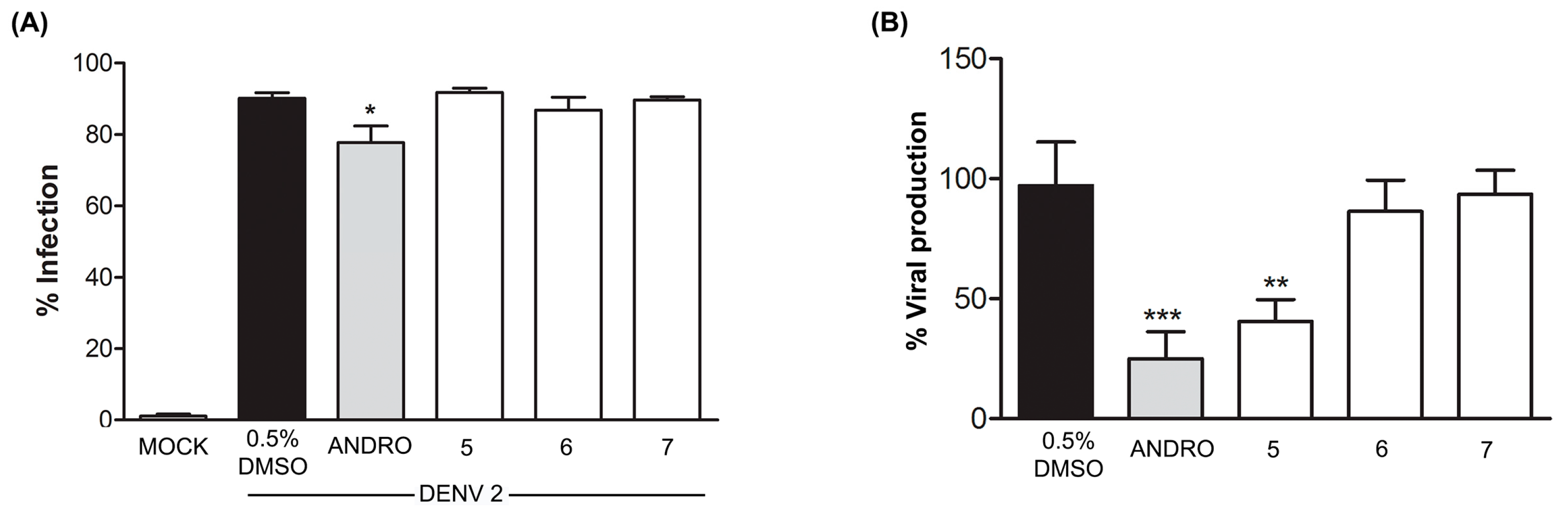
2.2.3. Effect of Compounds 5–7 on DENV Infection
3. Materials and Methods
3.1. General Procedures
3.2. Fungal Material
3.3. Culture, Extraction, and Isolation
3.4. Cell line and DENV2
3.5. Compounds Preparation
3.6. Cytotoxicity Assay
3.7. Antiviral Activity against DENV 2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kato, F.; Hishiki, T. Dengue virus reporter replicon is a valuable tool for antiviral drug discovery and analysis of virus replication mechanisms. Viruses 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Rivara, M.; Zuliani, V.; Radi, M. Drug repurposing approaches to fight dengue virus infection and related diseases. Front. Biosci. Landmark 2018, 23, 997–1019. [Google Scholar]
- Wadood, A.; Mehmood, A.; Khan, H.; Ilyas, M.; Ahmad, A.; Alarjan, M.; Abu-Izneid, T. Epitopes based drug design for dengue virus envelope protein: A computational approach. Comput. Biol. Chem. 2017, 71, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Morales, I.; Van Loock, M. An industry perspective on dengue drug discovery and development. Adv. Exp. Med. Biol. 2018, 1062, 333–353. [Google Scholar] [PubMed]
- Gerwick, W.H.; Fenner, A.M. Drug discovery from marine microbes. Microb. Ecol. 2013, 65, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.R.; Pereira, W.L.; Oliveira, A.F.C.D.; da Silva, A.M.; de Oliveira, A.S.; da Silva, M.L.; da Silva, C.C.; de Paula, S.O. Natural products as source of potential dengue antivirals. Molecules 2014, 19, 8151–8181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Raekiansyah, M.; Mori, M.; Nonaka, K.; Agoh, M.; Shiomi, K.; Matsumoto, A.; Morita, K. Identification of novel antiviral of fungus-derived brefeldin A against dengue viruses. Trop. Med. Int. Health 2017, 45, 32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.X.; Xu, M.Y.; Li, H.J.; Zeng, K.J.; Ma, W.Z.; Tian, G.B.; Xu, J.; Yang, D.P.; Lan, W.J. Diverse secondary metabolites from the marine-derived fungus Dichotomomyces cejpii F31-1. Mar. Drugs 2017, 15, 339. [Google Scholar] [CrossRef] [PubMed]
- Panraksa, P.; Ramphan, S.; Khongwichit, S.; Smith, D.R. Activity of andrographolide against dengue virus. Antivir. Res. 2017, 139, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Das, J.; Bhan, A.; Mandal, S.S.; Lovely, C.J. Total syntheses and cytotoxicity of kealiiquinone, 2-deoxy-2-aminokealiiquinone and analogs. Bioorg. Med. Chem. Lett. 2013, 23, 6183–6187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.H.; Xu, M.Y.; Li, H.J.; Li, J.Q.; Chen, Y.X.; Ma, W.Z.; Li, Y.P.; Xu, J.; Yang, D.P.; Lan, W.J. Aminoacid-directed strategy for inducing the marine-derived fungus Scedosporium apiospermum F41-1 to maximize alkaloid diversity. Org. Lett. 2017, 19, 4888–4891. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Inoue, Y.; Sekine, M.; Hayakawa, Y.; Homma, H.; Omura, S.; Tomoda, H. Relative and absolute stereochemistry of quinadoline B, an inhibitor of lipid droplet synthesis in macrophages. Org. Lett. 2008, 10, 5273–5276. [Google Scholar] [CrossRef] [PubMed]
- Sithisarn, P.; Suksanpaisan, L.; Thepparit, C.; Smith, D.R. Behavior of the dengue virus in solution. J. Med. Virol. 2003, 71, 532–539. [Google Scholar] [CrossRef] [PubMed]
| No. | 1 | |
|---|---|---|
| δC, Type | δH, Mult. (J, Hz) | |
| 1 | 145.3, C | |
| 2 | 133.4, C | |
| 3 | 96.5, CH | 7.25, s |
| 4 | 153.7, C | |
| 5 | 128.1, C | |
| 6 | 123.2, CH | 8.32, dd (7.2, 1.6) |
| 7 | 127.1, CH | 7.57, ddd (7.2, 7.2, 1.2) |
| 8 | 127.6, CH | 7.61, ddd (7.2, 7.2, 1.6) |
| 9 | 121.9, CH | 8.04, dd (7.2, 1.2) |
| 10 | 128.2, C | |
| 11 | 63.0, OCH3 | 3.98, s |
| 12 | 42.5, SOCH3 | 2.84, s |
| 13 | 56.3, OCH3 | 4.08, s |
| No. | 2 | 3 | 4 | |||
|---|---|---|---|---|---|---|
| δC, Type | δH, Mult. (J, Hz) | δC, Type | δH, Mult. (J, Hz) | δC, Type | δH, Mult. (J, Hz) | |
| 1 | 207.7, CO | 158.6, C | 116.6, C | |||
| 2 | 43.7, CH2 | 2.76, dd (14.8, 8.0) | 154.8, C | 128.7, C | ||
| 3 | 29.1, CH2 | 2.82, dd (14.0, 7.2) | 40.6, CH2 | 4.26, s | 128.4, C | |
| 4 | 133.0, C | 129.5, C | 104.3, CH | 7.24, s | ||
| 5 | 129.4, CH | 7.05, d (8.4) | 129.8, CH | 7.04, d (8,4) | 128.4, C | |
| 6 | 114.0, CH | 6.79, d (8.4) | 114.3, CH | 6.80, d (8.4) | 102.9, CH | 6.94, s |
| 7 | 158.1, C | 158.6, C | 151.7, C | |||
| 8 | 114.0, CH | 6.79, d (8.4) | 114.3, CH | 6.80, d (8.4) | 141.2, C | |
| 9 | 129.4, CH | 7.05, d (8.4) | 129.8, CH | 7.04, d (8,4) | 149.8, C | |
| 10 | 50.8, CH2 | 3.57, s | 41.2, CH2 | 4.10, s | 119.1, C | |
| 11 | 129.8, C | 132.9, C | 130.4, C | |||
| 12 | 106.5, CH | 6.34, s | 106.1, CH | 6.21, s | 130.3, CH | 7.29, d (8.4) |
| 13 | 153.5, C | 153.4, C | 113.6, CH | 7.00, d (8.4) | ||
| 14 | 137.1, C | 137.0, C | 158.8, CH | 7.24, s | ||
| 15 | 153.5, C | 153.4, C | 113.6, CH | 7.00, d (8.4) | ||
| 16 | 106.5, CH | 6.34, s | 106.1, CH | 6.22, s | 130.3, CH | 7.29, d (8.4) |
| 17 | 55.4, CH3 | 3.77, s | NH | 7.34, brs | ||
| 18 | 56.2, CH3 | 3.82, s | 143.7, CH | 9.15, s | 155.9, CO | |
| 19 | 61.0, CH3 | 3.83, s | 140.6, C | NH | 8.58, brs | |
| 20 | 56.2, CH3 | 3.82, s | 55.9, OCH3 | 3.96, s | ||
| 21 | 165.1, CO | 61.2, OCH3 | 3.88, s | |||
| 22 | 53.2, OCH3 | 4.04, s | 60.7, OCH3 55.5, OCH3 | 3.28, s 3.88, s | ||
| 23 | 55.4, OCH3 | 3.73, s | ||||
| 24 | 56.2, OCH3 | 3.74, s | ||||
| 25 | 61.0, OCH3 | 3.80, s | ||||
| 26 | 56.2, OCH3 | 3.73, s |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.-L.; Li, H.-J.; Smith, D.R.; Jaratsittisin, J.; Xia-Ke-Er, X.-F.-K.-T.; Ma, W.-Z.; Guo, Y.-W.; Dong, J.; Shen, J.; Yang, D.-P.; et al. Polyketides and Alkaloids from the Marine-Derived Fungus Dichotomomyces cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus. Mar. Drugs 2018, 16, 229. https://doi.org/10.3390/md16070229
Wu D-L, Li H-J, Smith DR, Jaratsittisin J, Xia-Ke-Er X-F-K-T, Ma W-Z, Guo Y-W, Dong J, Shen J, Yang D-P, et al. Polyketides and Alkaloids from the Marine-Derived Fungus Dichotomomyces cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus. Marine Drugs. 2018; 16(7):229. https://doi.org/10.3390/md16070229
Chicago/Turabian StyleWu, Dong-Lan, Hou-Jin Li, Duncan R. Smith, Janejira Jaratsittisin, Xia-Fu-Kai-Ti Xia-Ke-Er, Wen-Zhe Ma, Yong-Wei Guo, Jun Dong, Juan Shen, De-Po Yang, and et al. 2018. "Polyketides and Alkaloids from the Marine-Derived Fungus Dichotomomyces cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus" Marine Drugs 16, no. 7: 229. https://doi.org/10.3390/md16070229





