Identification and Characterization of Mycemycin Biosynthetic Gene Clusters in Streptomyces olivaceus FXJ8.012 and Streptomyces sp. FXJ1.235
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequence Analyses and Gene Organizations of the Mycemycin Biosynthetic Gene Clusters
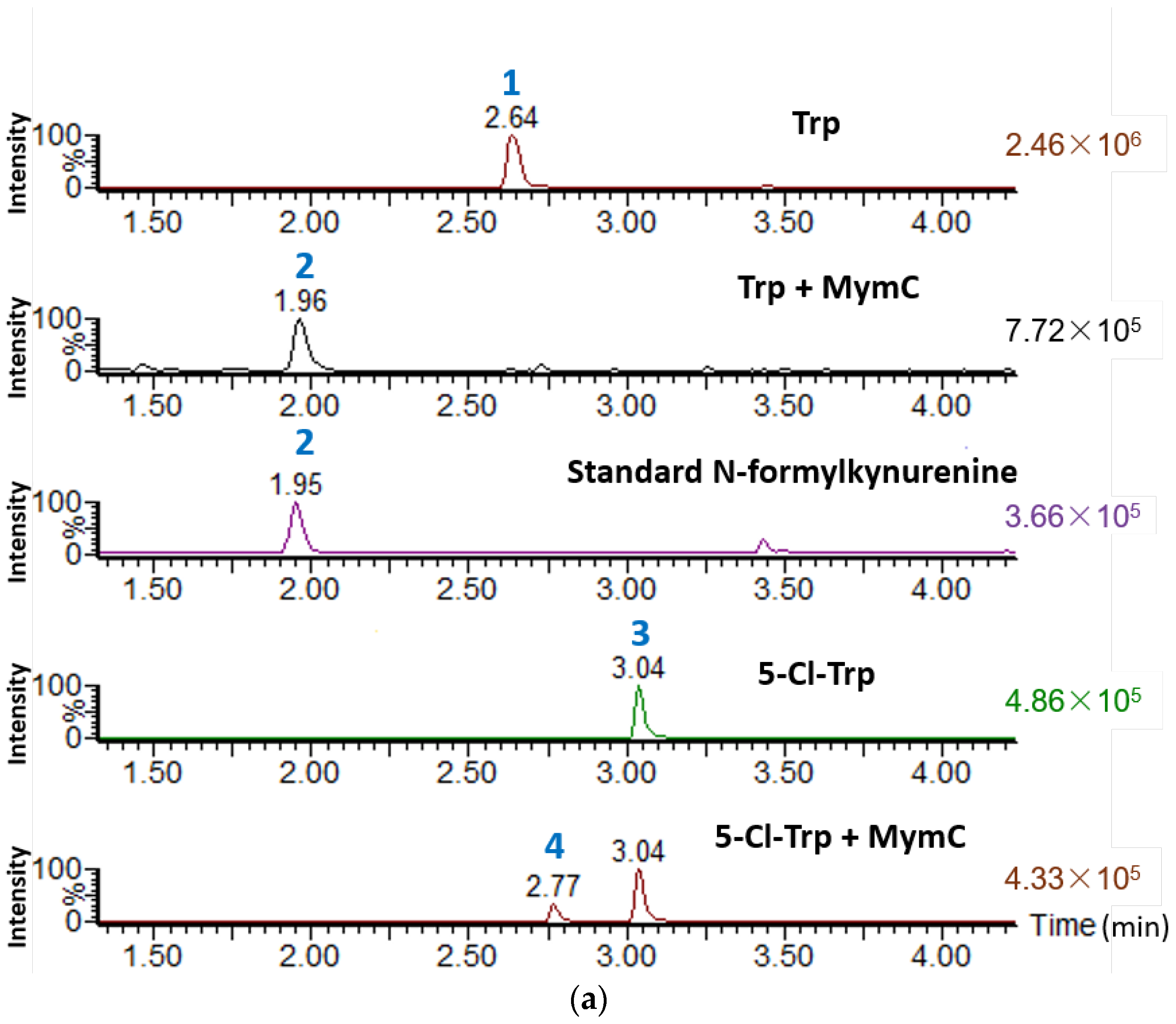
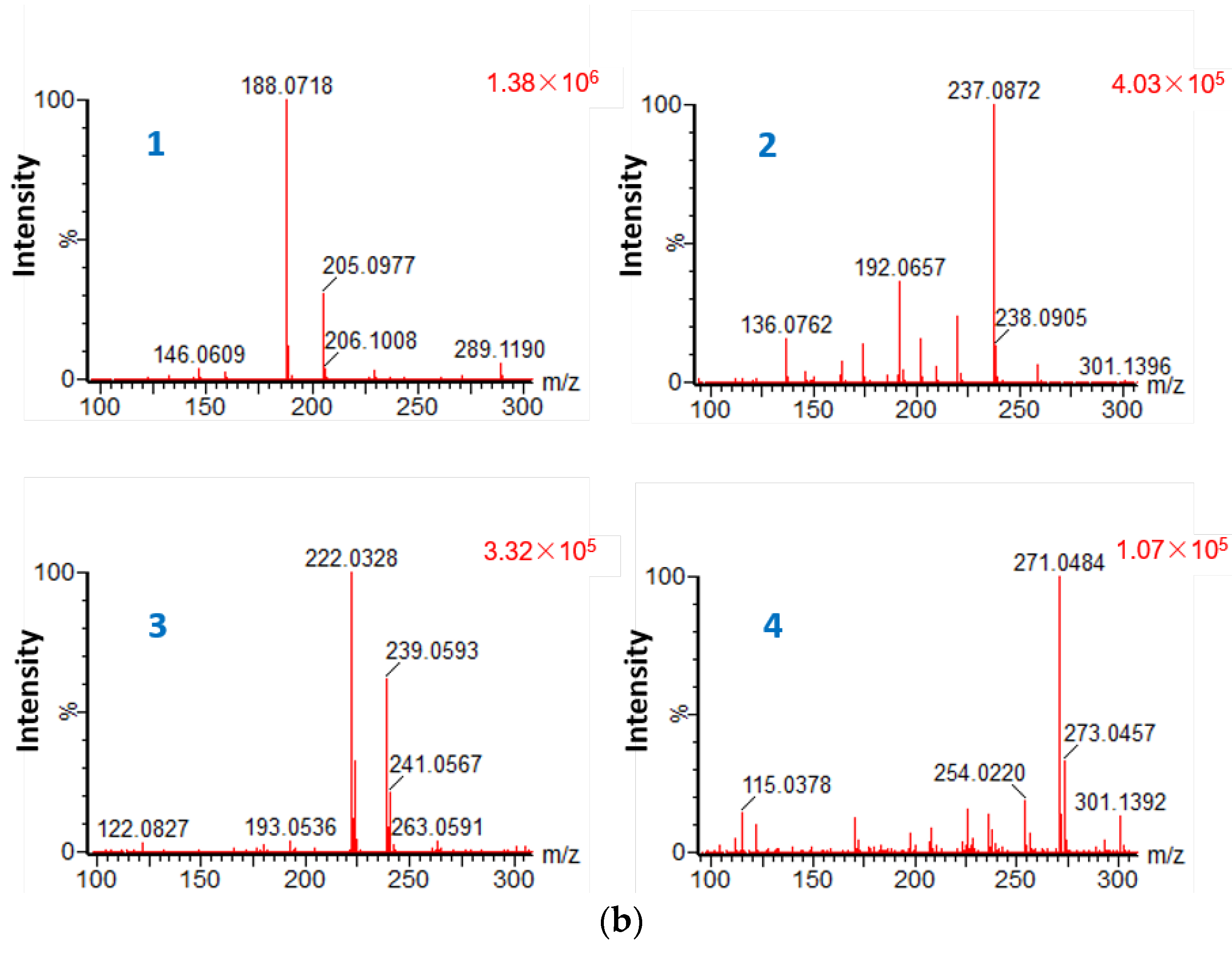
2.2. Heterologous Expression of the Key Enzyme Genes of Mym and in Vitro Activity Assay
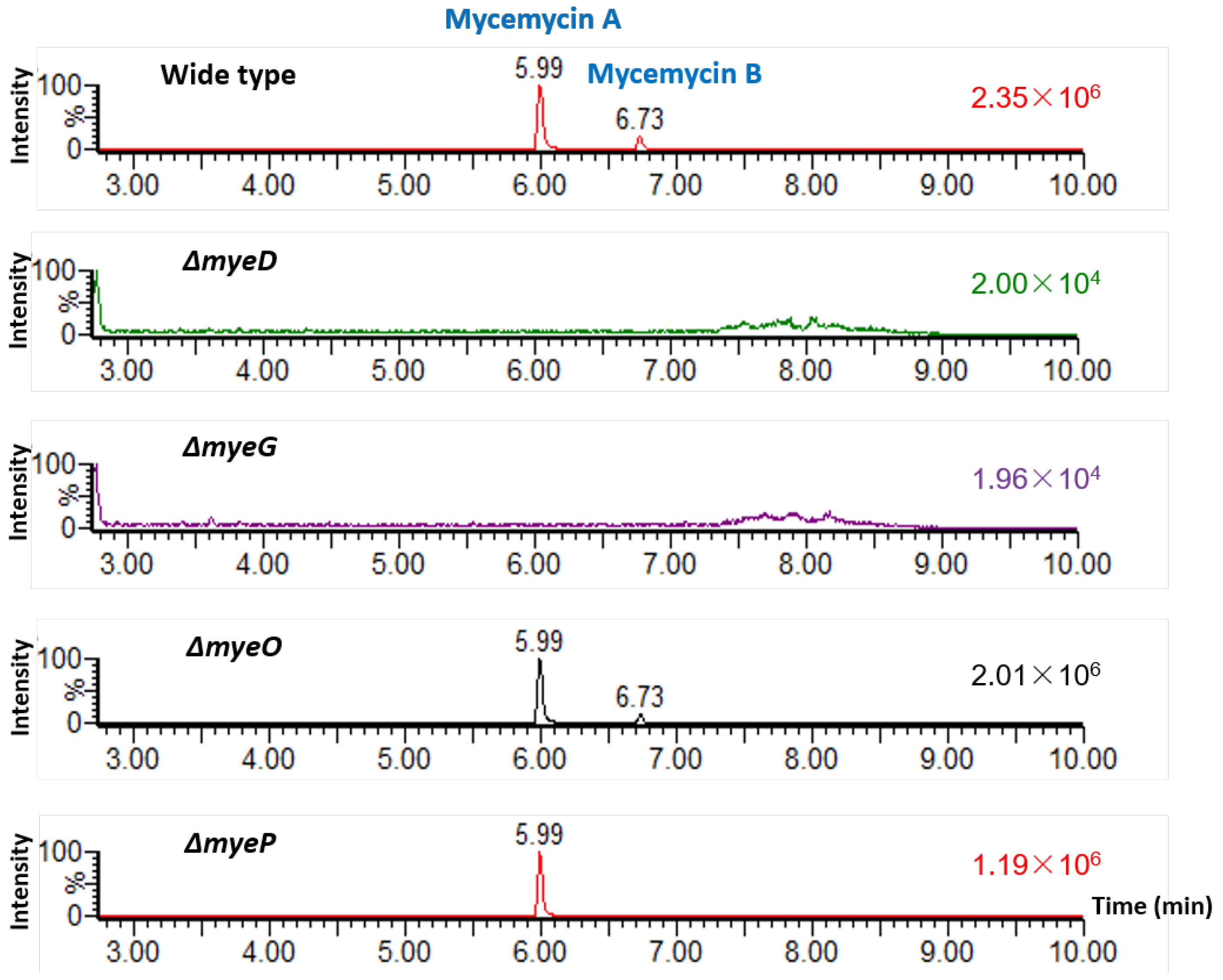
2.3. Identification of the Mycemycin Biosynthetic Gene Cluster in S. sp. FXJ1.235
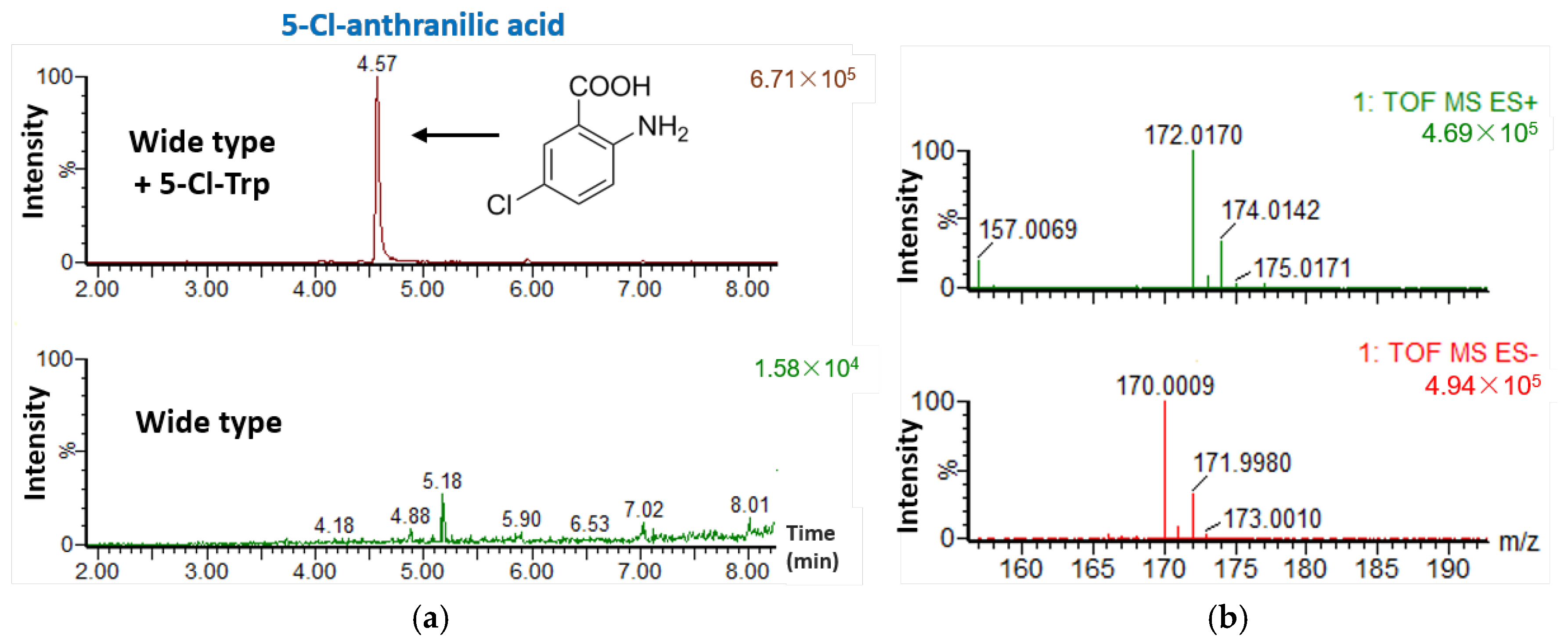
2.4. Feeding S. sp. FXJ1.235 with 5-Cl-Trp
2.5. Proposed Pathways for Mycemycin Biosynthesis
3. Materials and Methods
3.1. Strains, Plasmids and Media
3.2. Genome Sequencing and Annotation
3.3. Heterologous Expression and In Vitro Activity Assay of the TDO MymC
3.4. Mutant Construction and Confirmation
3.5. Feeding Experiments
3.6. Ultra-High-Performance Liquid Chromatography-High-Resolution Mass Spectrometry
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guo, X.L.; Zhang-Negrerie, D.; Du, Y.F. Iodine(III)-mediated construction of the dibenzoxazepinone skeleton from 2-(aryloxy) benzamides through oxidative C–N formation. RSC Adv. 2015, 5, 94732–94736. [Google Scholar] [CrossRef]
- Fiorentino, A.; D’Abrosca, B.; Pacifico, S.; Cefarelli, G.; Uzzo, P.; Monaco, P. Natural dibenzoxazepinones from leaves of Carex distachya: Structural elucidation and radical scavenging activity. Bioorg. Med. Chem. Lett. 2007, 17, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Peng, Q.; Zhang, R.; Hu, J.; Zhou, Y.; Xu, L.; Pan, X. Ligand-controlled chemoselective one-pot synthesis of dibenzo-thiazepinones and dibenzoxazepinones via twice copper-catalyzed cross-coupling. Synlett 2017, 28, 1201–1208. [Google Scholar]
- Zhou, Y.; Zhu, J.; Li, B.; Zhang, Y.; Feng, J.; Hall, A.; Shi, J.; Zhu, W. Access to different isomeric dibenzoxazepinones through copper-catalyzed C–H etherification and C–N bond construction with controllable Smiles rearrangement. Org. Lett. 2016, 18, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, Y.; Qian, P.; Hu, Y.; Ao, G.; Chen, S.; Zhang, S.; Zhang, Y. An efficient cascade approach to dibenzoxazepinones via nucleophilic aromatic substitution and Smiles rearrangement. Tetrahedron Lett. 2015, 56, 2211–2213. [Google Scholar] [CrossRef]
- Hone, N.D.; Salter, J.I.; Reader, J.C. Solid-phase synthesis of dibenzoxazepinones. Tetrahedron Lett. 2003, 44, 8169–8172. [Google Scholar] [CrossRef]
- Liu, N.; Song, F.; Shang, F.; Huang, Y. Mycemycins A–E, new dibenzoxazepinones isolated from two different Streptomycetes. Mar. Drugs 2015, 13, 6247–6258. [Google Scholar] [CrossRef] [PubMed]
- Sono, M.; Roach, M.P.; Coulter, E.D.; Dawson, J.H. Heme-containing oxygenases. Chem. Rev. 1996, 96, 2841–2888. [Google Scholar] [CrossRef] [PubMed]
- Forouhar, F.; Anderson, J.L.R.; Mowat, C.G.; Vorobiev, S.M.; Hussain, A.; Abashidze, M.; Bruckmann, C.; Thackray, S.J.; Seetharaman, J.; Tucker, T.; et al. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 2007, 104, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Kurnasov, O.; Goral, V.; Colabroy, K.; Gerdes, S.; Anantha, S.; Osterman, A.; Begley, T.P. NAD biosynthesis: Identification of the tryptophan to quinolinate pathway in Bacteria. Chem. Biol. 2003, 10, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Phelan, V.; Ntai, I.; Farnet, C.M.; Zazopoulos, E.; Bachmann, B.O. Benzodiazepine biosynthesis in Streptomyces refuineus. Chem. Biol. 2007, 14, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Keller, U.; Lang, M.; Crnovcic, I.; Pfennig, F.; Schauwecker, F. The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: A genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 2010, 192, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, S.; Baysse, C.; Koedam, N.; Tehrani, K.A.; Verheyden, L.; Budzikiewicz, H.; Schafer, M.; Hoorelbeke, B.; Meyer, J.M.; De Greve, H.; et al. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 2004, 52, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Khullar, A.; Chou, S.; Sacramo, A.; Gerratana, B. Biosynthesis of sibiromycin, a potent antitumor antibiotic. Appl. Environ. Microbiol. 2009, 75, 2869–2878. [Google Scholar] [CrossRef] [PubMed]
- Batabyal, D.; Yeh, S.-R. Human tryptophan dioxygenase: A comparison to indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2007, 129, 15690–15701. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, D.G.; Hrvatin, S.; Neumann, C.S.; Strieker, M.; Marahiel, M.A.; Walsh, C.T. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc. Natl. Acad. Sci. USA 2007, 104, 16498–16503. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.; Garneau, S.; Walsh, C.T. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 3960–3965. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.R.; Chapman, S.K. Molecular mechanisms of enzyme-catalysed halogenation. Mol. Biosyst. 2006, 2, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Milbredt, D.; Patallo, E.P.; van Pée, K.-H. A tryptophan 6-halogenase and an amidotransferase are involved in thienodolin biosynthesis. ChemBioChem 2014, 15, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Serino, L.; Reimmann, C.; Baur, H.; Beyeler, M.; Visca, P.; Haas, D. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 1995, 249, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Serino, L.; Reimmann, C.; Visca, P.; Beyeler, M.; Chiesa, V.D.; Haas, D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhao, J.; Deng, Z.; Yu, Y. Characterization of the biosynthetic gene cluster for benzoxazole antibiotics A33853 reveals unusual assembly logic. Chem. Biol. 2015, 22, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Blanton, J.M.; Podell, S.; Taton, A.; Schorn, M.A.; Busch, J.; Lin, Z.; Schmidt, E.W.; Jensen, P.R.; Paul, V.J.; et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 2017, 13, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; El Gamal, A.A.; Yamanaka, K.; Poth, D.; Kersten, R.D.; Schorn, M.; Allen, E.E.; Moore, B.S. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 2014, 10, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, O.; Luzhetskyy, A. Metabolic engineering of natural product biosynthesis in actinobacteria. Curr. Opin. Biotechnol. 2016, 42, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Knoten, C.A.; Hudson, L.L.; Coleman, J.P.; Farrow, J.M.; Pesci, E.C. KynR, a Lrp/AsnC-type transcriptional regulator, directly controls the kynurenine pathway in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 6567–6575. [Google Scholar] [CrossRef] [PubMed]
- Zummo, F.P.; Marineo, S.; Pace, A.; Civiletti, F.; Giardina, A.; Puglia, A.M. Tryptophan catabolism via kynurenine production in Streptomyces coelicolor: Identification of three genes coding for the enzymes of tryptophan to anthranilate pathway. Appl. Microbiol. Biotechnol. 2012, 94, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]



| Gene | Size (AA) | Protein Homolog and Origin (Identity/Similarity) | Homolog in mye (Identity/Similarity) | Proposed Function |
|---|---|---|---|---|
| orf-1 | 231 | WP_011030904.1 (89/95); Streptomyces coelicolor | - | Hypothetical protein |
| orf-2 | 78 | None predicted in NCBI | - | Unknown |
| mymA | 454 | SsfH, ADE34507.1 (55/68); Streptomyces sp. SF2575 | myeA (53/66) | Salicylate synthase |
| mymB | 351 | SibL, ACN39735.1 (65/76); Streptosporangium sibiricum | - | Methyltransferase |
| mymC | 269 | SibP, ACN39739.1 (59/70); Streptosporangium sibiricum | myeC (38/53) | Tryptophan 2,3-dioxygenase (TDO) |
| mymD | 513 | EFL38513.1 (61/71); Streptomyces griseoflavus Tu4000 | myeD (62/76) | Amidohydrolase |
| mymE | 82 | EsmD3, AFB35628.1 (48/61); Streptomyces antibioticus | myeE (49/63) | Phosphopantetheine-binding protein |
| mymF | 547 | PchD, NP_252918.1 (48/61); Pseudomonas aeruginosa PAO1 | myeF (61/73) | 2,3-dihydroxybenzoate-AMP ligase |
| mymG | 339 | BomK, ALE27503.1 (52/70); Streptomyces sp. NRRL 12068 | myeG (60/76) | Beta-ketoacyl-ACP synthase (amide bond formation) |
| mymH | 220 | EFL35401.1 (49/64); Streptomyces viridochromogenes DSM 40736 | - | Hypothetical protein |
| mymI | 354 | KUK37103.1 (61/75); Thermacetogenium phaeum | - | 3-deoxy-7-phosphoheptulonate synthase |
| mymJ | 281 | SCE38941.1 (60/68); Streptomyces sp. PpalLS-921 | myeJ (63/72) | SAM-dependent methyltransferase |
| mymR1 | 329 | SBU95411.1 (75/86); Streptomyces sp. OspMP-M45 | myeR1 (75/85) | Lrp/AsnC family transcriptional regulator |
| mymK | 563 | CB02009_orf6, OKJ63402.1 (69/81); Streptomyces sp. CB02009 | myeK (69/79) | Multidrug MFS transporter |
| mymL | 294 | Carboxylesterase NlhH, ASO22469.1 (50/61); Actinoalloteichus hoggarensis | - | Alpha/beta hydrolase |
| mymR2 | 1072 | WP_033441976.1 (38/49); Saccharothrix sp. NRRL B-16314 | - | SARP family transcriptional regulator |
| mymM | 511 | PyrH, OSY47217.1 (61/76); Streptomyces platensis | - | Tryptophan halogenase |
| mymN | 464 | SDU28343.1 (53/68); Amycolatopsis keratiniphila | myeN (61/74, query cover 56%) | Sodium/hydrogen exchanger family |
| mymO | 175 | KtzS, ABV56599.1 (59/67); Kutzneria sp. 744 | myeO (61/69) | Flavin reductase |
| mymP | 530 | RebH, 4LU6_A (63/76); Lechevalieria aerocolonigenes | myeP (75/83) | Tryptophan halogenase |
| mymQ | 411 | KynU, NP_250770.1 (44/60); Pseudomonas aeruginosa PAO1 | myeQ (51/64) | Kynureninase (KYN) |
| mymR3 | 691 | SBU95407.1 (56/67); Streptomyces sp. OspMP-M45 | myeR3 (57/68) | SARP family transcriptional regulator |
| orf-3 | 38 | None predicted in NCBI | - | Unknown |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, F.; Liu, N.; Liu, M.; Chen, Y.; Huang, Y. Identification and Characterization of Mycemycin Biosynthetic Gene Clusters in Streptomyces olivaceus FXJ8.012 and Streptomyces sp. FXJ1.235. Mar. Drugs 2018, 16, 98. https://doi.org/10.3390/md16030098
Song F, Liu N, Liu M, Chen Y, Huang Y. Identification and Characterization of Mycemycin Biosynthetic Gene Clusters in Streptomyces olivaceus FXJ8.012 and Streptomyces sp. FXJ1.235. Marine Drugs. 2018; 16(3):98. https://doi.org/10.3390/md16030098
Chicago/Turabian StyleSong, Fangying, Ning Liu, Minghao Liu, Yihua Chen, and Ying Huang. 2018. "Identification and Characterization of Mycemycin Biosynthetic Gene Clusters in Streptomyces olivaceus FXJ8.012 and Streptomyces sp. FXJ1.235" Marine Drugs 16, no. 3: 98. https://doi.org/10.3390/md16030098





