Novel Antimicrobial Peptides from the Arctic Polychaeta Nicomache minor Provide New Molecular Insight into Biological Role of the BRICHOS Domain
Abstract
:1. Introduction
2. Results and Discussion
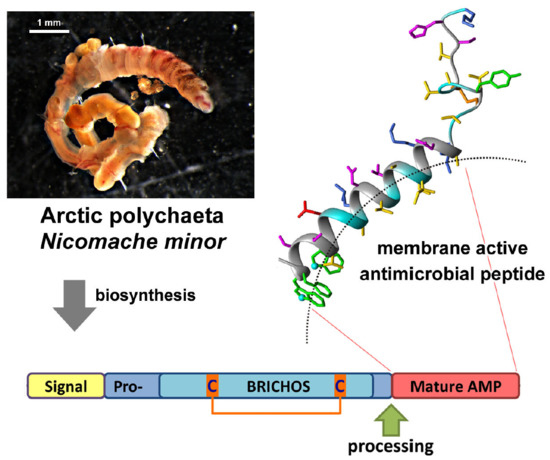
2.1. Nicomicin Is a Novel BRICHOS Domain-Related AMP
2.2. Nicomicin Is Unique Among Polychaeta AMPs but Shares Structural Similarities with Other Animal Host-Defense Peptides
2.3. Recombinant Expression and Purification of Nicomicin-1 and Its Fragments
2.4. Nicomicin-1 Is Disordered in Aqueous Solution but Forms an α-Helical Structure in a Membrane-Mimicking Environment
2.5. Topology of Nicomicin-Micelle Interaction
2.6. Comparison of Spacial Structures of Nicomicin-1 and Amphibian Peptides Containing ‘Rana-Box’ Motif
2.7. Antibacterial Activity and Mechanism of Action
2.8. Nicomicin-1 Possesses Cytotoxicity Against Mammalian Cells
3. Materials and Methods
3.1. Animal Collection
3.2. Total RNA Isolation, RT-PCR, RACE Amplification
3.3. Expression and Purification of the Antimicrobial Peptides
3.4. Circular Dichroism Spectroscopy
3.5. NMR Spectroscopy
3.6. PDB and BMRB Accession Codes
3.7. Tryptophan Fluorescence and Quenching
3.8. Antimicrobial Assay
3.9. Bacterial Membranes Permeability Assay
3.10. Cell-Free Protein Expression Assay
3.11. Hemolysis and Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hancock, R.E.W.; Brown, K.L.; Mookherjee, N. Host defence peptides from invertebrates—Emerging antimicrobial strategies. Immunobiology 2006, 211, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Mardirossian, M.; Nguyen, F.; Seefeldt, A.C.; Guichard, G.; Scocchi, M.; Innis, C.A.; Wilson, D.N. Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 2017, 34, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Tasiemski, A. Antimicrobial peptides in annelids. Invertebr. Surviv. J. 2008, 5, 75–82. [Google Scholar]
- Ovchinnikova, T.V.; Aleshina, G.M.; Balandin, S.V.; Krasnosdembskaya, A.D.; Markelov, M.L.; Frolova, E.I.; Leonova, Y.F.; Tagaev, A.A.; Krasnodembsky, E.G.; Kokryakov, V.N. Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 2004, 577, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Shenkarev, Z.O.; Nadezhdin, K.D.; Balandin, S.V.; Zhmak, M.N.; Kudelina, I.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S. Recombinant expression, synthesis, purification, and solution structure of arenicin. Biochem. Biophys. Res. Commun. 2007, 360, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Liu, X.; Ge, F.; Han, J.; Zheng, T. Perinerin, a novel antimicrobial peptide purified from the clamworm Perinereis aibuhitensis Grube and its partial characterization. J. Biochem. 2004, 135, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Tasiemski, A.; Schikorski, D.; Le Marrec-Croq, F.; Pontoire-Van Camp, C.; Boidin-Wichlacz, C.; Sautière, P.-E. Hedistin: A novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid, Nereis diversicolor. Dev. Comp. Immunol. 2007, 31, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Tasiemski, A.; Jung, S.; Boidin-Wichlacz, C.; Jollivet, D.; Cuvillier-Hot, V.; Pradillon, F.; Vetriani, C.; Hecht, O.; Sönnichsen, F.D.; Gelhaus, C.; et al. Characterization and function of the first antibiotic isolated from a vent organism: The extremophile metazoan Alvinella pompejana. PLoS ONE 2014, 9, e95737. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, T.D.; Tzetlin, A.B.; Mardashova, M.V.; Sokolova, O.S. Fine structure of the tubes of Maldanidae (Annelida). J. Mar. Biol. Assoc. U. K. 2017, 97, 1177–1187. [Google Scholar] [CrossRef]
- Chen, G.; Abelein, A.; Nilsson, H.E.; Leppert, A.; Andrade-Talavera, Y.; Tambaro, S.; Hemmingsson, L.; Roshan, F.; Landreh, M.; Biverstål, H.; et al. Bri2 BRICHOS client specificity and chaperone activity are governed by assembly state. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Leonard, B.C.; Chu, H.; Johns, J.L.; Gallo, R.L.; Moore, P.F.; Marks, S.L.; Bevins, C.L. Expression and activity of a novel cathelicidin from domestic cats. PLoS ONE 2011, 6, e18756. [Google Scholar] [CrossRef] [PubMed]
- Hoegenhaug, H.H.K.; Mygind, P.H.; Kruse, T.; Segura, D.R.; Sandvang, D.; Neve, S. Antimicrobial Peptide Variants and Polynucleotides Encoding Same. US Patent US20110306750A1; PCT/EP2011/059689, 15 December 2011. [Google Scholar]
- Papot, C.; Massol, F.; Jollivet, D.; Tasiemski, A. Antagonistic evolution of an antibiotic and its molecular chaperone: How to maintain a vital ectosymbiosis in a highly fluctuating habitat. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Bergeron, F.; Leduc, R.; Day, R. Subtilase-like pro-protein convertases: From molecular specificity to therapeutic applications. J. Mol. Endocrinol. 2000, 24, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Nitta, H.; Wada, Y.; Kobayashi, H.; Okamoto, K. Impaired plasma clottability induction through fibrinogen degradation by ASP, a serine protease released from Aeromonas sobria. FEMS Microbiol. Lett. 2008, 284, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, J.; Johansson, J.; Persson, B. BRICHOS—A superfamily of multidomain proteins with diverse functions. BMC Res. Notes 2009, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.D.; Bornberg-Bauer, E. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 2012, 29, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Song, K.; Kim, S.; Lee, J.; Hwang, S.; Han, C. Caenorhabditis elegans BRICHOS domain–containing protein C09F5.1 maintains thermotolerance and decreases cytotoxicity of Aβ42 by activating the UPR. Genes 2018, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pulido, L.; Devos, D.; Valencia, A. BRICHOS: A conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem. Sci. 2002, 27, 329–332. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Li, X.; Yang, X.; Yu, X.; Wang, J.; Liu, F.; Huang, D. Transcriptional response of Musca domestica larvae to bacterial infection. PLoS ONE 2014, 9, e104867. [Google Scholar] [CrossRef] [PubMed]
- Tomie, T.; Ishibashi, J.; Furukawa, S.; Kobayashi, S.; Sawahata, R.; Asaoka, A.; Tagawa, M.; Yamakawa, M. Scarabaecin, a novel cysteine-containing antifungal peptide from the rhinoceros beetle, Oryctes rhinoceros. Biochem. Biophys. Res. Commun. 2003, 307, 261–266. [Google Scholar] [CrossRef]
- Yang, H.; Wang, X.; Liu, X.; Wu, J.; Liu, C.; Gong, W.; Zhao, Z.; Hong, J.; Lin, D.; Wang, Y.; et al. Antioxidant peptidomics reveals novel skin antioxidant system. Mol. Cell. Proteom. 2009, 8, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Basir, Y.J.; Knoop, F.C.; Dulka, J.; Conlon, J.M. Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim. Biophys. Acta 2000, 1543, 95–105. [Google Scholar] [CrossRef]
- Haney, E.F.; Hunter, H.N.; Matsuzaki, K.; Vogel, H.J. Solution NMR studies of amphibian antimicrobial peptides: Linking structure to function? Biochim. Biophys. Acta Biomembr. 2009, 1788, 1639–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubovskii, P.V.; Vassilevski, A.A.; Samsonova, O.V.; Egorova, N.S.; Kozlov, S.A.; Feofanov, A.V.; Arseniev, A.S.; Grishin, E.V. Novel lynx spider toxin shares common molecular architecture with defense peptides from frog skin: Spider toxin with a single disulfide bond. FEBS J. 2011, 278, 4382–4393. [Google Scholar] [CrossRef] [PubMed]
- Haugen, H.S.; Fimland, G.; Nissen-Meyer, J.; Kristiansen, P.E. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A. Biochemistry 2005, 44, 16149–16157. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.M.; Xu, S.; Sheftic, S.R.; Alexandrescu, A.T. Dynamic α-helix structure of micelle-bound human amylin. J. Biol. Chem. 2009, 284, 11982–11991. [Google Scholar] [CrossRef] [PubMed]
- Nanga, R.P.R.; Brender, J.R.; Vivekanandan, S.; Ramamoorthy, A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Q.; Chen, J.-C.; Cui, Y.-X.; Zhou, B.; Chen, Y.-X.; Zhao, Y.-F.; Li, Y.-M. Antimicrobial activity of human islet amyloid polypeptides: An insight into amyloid peptides’ connection with antimicrobial peptides. Biol. Chem. 2012, 393. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, M.E.; Hermansson, E.; Wang, Y.; Welsh, N.; Presto, J.; Johansson, J.; Westermark, G.T. BRICHOS domain of Bri2 inhibits islet amyloid polypeptide (IAPP) fibril formation and toxicity in human beta cells. Proc. Natl. Acad. Sci. USA 2018, 115, E2752–E2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.; Cao, L.; Ma, Y.; Mao, P.; Wang, W.; Zhao, R.; Wu, Y.; Cao, Z.; Li, W. Cloning and functional characterization of a new antimicrobial peptide gene StCT1 from the venom of the scorpion Scorpiops tibetanus. Peptides 2010, 31, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Vargas-Jaimes, L.; Batista, C.V.F.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Aleinein, R.A.; Hamoud, R.; Schäfer, H.; Wink, M. Molecular cloning and expression of ranalexin, a bioactive antimicrobial peptide from Rana catesbeiana in Escherichia coli and assessments of its biological activities. Appl. Microbiol. Biotechnol. 2013, 97, 3535–3543. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, Q.; Li, Z.; Zhang, Y.; Zhao, J.; Wang, L. Molecular cloning, expression, purification, and functional characterization of palustrin-2CE, an antimicrobial peptide of Rana chensinensis. Biosci. Biotechnol. Biochem. 2012, 76, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, M.; Li, C. Cloning and expression of a novel insulin-releasing peptide, brevinin-2GU from Escherichia coli. J. Biosci. Bioeng. 2009, 107, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Shenkarev, Z.O.; Balandin, S.V.; Nadezhdin, K.D.; Paramonov, A.S.; Kokryakov, V.N.; Arseniev, A.S. Molecular insight into mechanism of antimicrobial action of the beta-hairpin peptide arenicin: Specific oligomerization in detergent micelles. Biopolymers 2008, 89, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Jean, L.; Lee, C.F.; Hodder, P.; Hawkins, N.; Vaux, D.J. Dynamics of the formation of a hydrogel by a pathogenic amyloid peptide: Islet amyloid polypeptide. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, A.N.; Liu, Y.; Wang, T.; Musgrave, I.F.; Pukala, T.L.; Tabor, R.F.; Martin, L.L.; Carver, J.A.; Bowie, J.H. The amyloid fibril-forming properties of the amphibian antimicrobial peptide uperin 3.5. ChemBioChem 2016, 17, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1957–1974. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, G.; Kumar, T.K.; Arunkumar, A.I.; Yu, C. 2,2,2-Trifluoroethanol induces helical conformation in an all beta-sheet protein. Biochem. Biophys. Res. Commun. 1996, 222, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G., III; Rance, M.; Skelton, N.J. Protein NMR Spectroscopy, 2nd ed.; Academic Press: Cambridge, MA, USA, 2006; ISBN 978-0-12164-491-8. [Google Scholar]
- Brown, L.R.; Bösch, C.; Wüthrich, K. Location and orientation relative to the micelle surface for glucagon in mixed micelles with dodecylphosphocholine: EPR and NMR studies. Biochim. Biophys. Acta 1981, 642, 296–312. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Szabo, A.G. Fluorescence principles and measurements. In Spectrophotometry and Spectrofluorimetry, 2nd ed.; Gore, M.G., Ed.; Oxford University Press: New York, NY, USA, 2000; pp. 33–67. ISBN 978-0-19963-812-3. [Google Scholar]
- Chi, S.-W.; Kim, J.-S.; Kim, D.-H.; Lee, S.-H.; Park, Y.-H.; Han, K.-H. Solution structure and membrane interaction mode of an antimicrobial peptide gaegurin 4. Biochem. Biophys. Res. Commun. 2007, 352, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Subasinghage, A.P.; Conlon, J.M.; Hewage, C.M. Conformational analysis of the broad-spectrum antibacterial peptide, ranatuerin-2CSa: Identification of a full length helix–turn–helix motif. Biochim. Biophys. Acta Proteins Proteom. 2008, 1784, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, S.; Nakagawa, K.; Maeno, M.; Momose, H. In vivo monitoring system for structure-function relationship analysis of the antibacterial peptide apidaecin. Appl. Environ. Microbiol. 1994, 60, 3566–3572. [Google Scholar] [PubMed]
- Yonezawa, A.; Kuwahara, J.; Fujii, N.; Sugiura, Y. Binding of tachyplesin I to DNA revealed by footprinting analysis: Significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 1992, 31, 2998–3004. [Google Scholar] [CrossRef] [PubMed]
- Maltseva, A.L.; Kotenko, O.N.; Kokryakov, V.N.; Starunov, V.V.; Krasnodembskaya, A.D. Expression pattern of arenicins—The antimicrobial peptides of polychaete Arenicola marina. Front. Physiol. 2014, 5, 497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Ovchinnikova, T.V. Improved strategy for recombinant production and purification of antimicrobial peptide tachyplesin I and its analogs with high cell selectivity. Biotechnol. Appl. Biochem. 2017, 64, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Bolosov, I.A.; Balandin, S.V.; Ovchinnikova, T.V. Design of antimicrobial peptide arenicin analogs with improved therapeutic indices. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2015, 21, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panteleev, P.V.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dimerization of the antimicrobial peptide arenicin plays a key role in the cytotoxicity but not in the antibacterial activity. Biochem. Biophys. Res. Commun. 2017, 482, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P. Automated NMR structure calculation with CYANA. In Protein NMR Techniques; Humana Press: Totowa, NJ, USA, 2004; Volume 278, pp. 353–378. ISBN 978-1-59259-809-0. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Shamova, O.V.; Orlov, D.S.; Zharkova, M.S.; Balandin, S.V.; Yamschikova, E.V.; Knappe, D.; Hoffmann, R.; Kokryakov, V.N.; Ovchinnikova, T.V. Minibactenecins ChBac7.Nα and ChBac7.Nβ—Antimicrobial peptides from leukocytes of the goat Capra hircus. Acta Nat. 2016, 8, 136–146. [Google Scholar]
- Kuzmin, D.V.; Emelianova, A.A.; Kalashnikova, M.B.; Panteleev, P.V.; Balandin, S.V.; Serebrovskaya, E.O.; Belogurova-Ovchinnikova, O.Y.; Ovchinnikova, T.V. Comparative in vitro study on cytotoxicity of recombinant β-hairpin peptides. Chem. Biol. Drug Des. 2018, 91, 294–303. [Google Scholar] [CrossRef] [PubMed]









| Primer Name | Sequence 5′→3′ |
|---|---|
| 3′-GSP1 | GTGTTACGTCATGGGTGG(G,C)(G,C)T(G,T)GAC |
| 3′-GSP2 | GAGTGCTAC(T,C)TG(A,G)TCGG(A,C)GG |
| 3′-GSP3 | TGC(G,T,C)AGGG(A,C)AA(A,G)CCTGT(C,T)TTCTGG(A,C)T |
| 5′-GSP1 | GTGGTCAATGAATATCTGCAATACA |
| 5′-GSP2 | GAGCTTATACCCATAGGGCTTCCTTATAC |
| 5′-GSP3 | ATTAAGAACGTTGTCCAAAGCGTAATG |
| AP | GTTGATCCGACAGTCGCTTGC |
| Peptide | RP-HPLC Retention Time 1 (min) | Calculated [M + H]+ Monoisotopic Mass (Da) | Measured Monoisotopic m/z Value 2 | Recombinant Peptide Final Yield (mg/L) |
|---|---|---|---|---|
| Nicomicin-1 | 63 | 3535.81 * | 3535.64 | 0.9 |
| Nico(1-17) | 56 | 1794.87 | 1794.84 | 6.2 |
| Nico(18-33) | 42.5 | 1759.94 * | 1759.97 | 4.3 |
| Bacteria | Minimum Inhibitory Concentration (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Melittin | Nicomicin-1 | Nico(1-17) | Nico(18-33) | |||||
| Without Salt | +NaCl | Without Salt | +NaCl | Without Salt | +NaCl | Without Salt | +NaCl | |
| Gram-positive | ||||||||
| Micrococcus luteus | 0.25 | 0.25 | 0.125 | 0.25 | >16 | >16 | 16 | >16 |
| Bacillus subtilis | 0.5 | 0.5 | 0.062 | 0.25 | >16 | >16 | 16 | >16 |
| B. licheniformis | 0.25 | 0.25 | 0.125 | 0.25 | >16 | >16 | 8 | >128 |
| B. megaterium | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Staphylococcus aureus 209P | 2 | 32 | 2 | 32 | >16 | >16 | >16 | >16 |
| S. aureus ATCC 29213 | 1 | 1 | 2 | 16 | >16 | >16 | >128 | >128 |
| Rhodococcus sp. | 0.5 | 0.25 | 0.125 | 0.25 | >16 | >16 | >16 | >16 |
| Gram-negative | ||||||||
| E. coli BL21 (DE3) | 2 | 4 | 2 | 32 | >64 | >64 | >64 | >64 |
| E. coli ML-35p | 8 | 16 | 16 | >32 | >64 | >64 | >64 | >64 |
| E. coli C600 | 4 | 8 | 32 | >32 | >64 | >64 | >64 | >64 |
| Acinetobacter baumanii | 8 | 32 | 32 | >32 | >64 | >64 | >64 | >64 |
| Pseudomonas aeruginosa PAO1 | >32 | 32 | 32 | >32 | >64 | >64 | >128 | >128 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panteleev, P.V.; Tsarev, A.V.; Bolosov, I.A.; Paramonov, A.S.; Marggraf, M.B.; Sychev, S.V.; Shenkarev, Z.O.; Ovchinnikova, T.V. Novel Antimicrobial Peptides from the Arctic Polychaeta Nicomache minor Provide New Molecular Insight into Biological Role of the BRICHOS Domain. Mar. Drugs 2018, 16, 401. https://doi.org/10.3390/md16110401
Panteleev PV, Tsarev AV, Bolosov IA, Paramonov AS, Marggraf MB, Sychev SV, Shenkarev ZO, Ovchinnikova TV. Novel Antimicrobial Peptides from the Arctic Polychaeta Nicomache minor Provide New Molecular Insight into Biological Role of the BRICHOS Domain. Marine Drugs. 2018; 16(11):401. https://doi.org/10.3390/md16110401
Chicago/Turabian StylePanteleev, Pavel V., Andrey V. Tsarev, Ilia A. Bolosov, Alexander S. Paramonov, Mariana B. Marggraf, Sergey V. Sychev, Zakhar O. Shenkarev, and Tatiana V. Ovchinnikova. 2018. "Novel Antimicrobial Peptides from the Arctic Polychaeta Nicomache minor Provide New Molecular Insight into Biological Role of the BRICHOS Domain" Marine Drugs 16, no. 11: 401. https://doi.org/10.3390/md16110401