Brevianamides and Mycophenolic Acid Derivatives from the Deep-Sea-Derived Fungus Penicillium brevicompactum DFFSCS025
Abstract
:1. Introduction
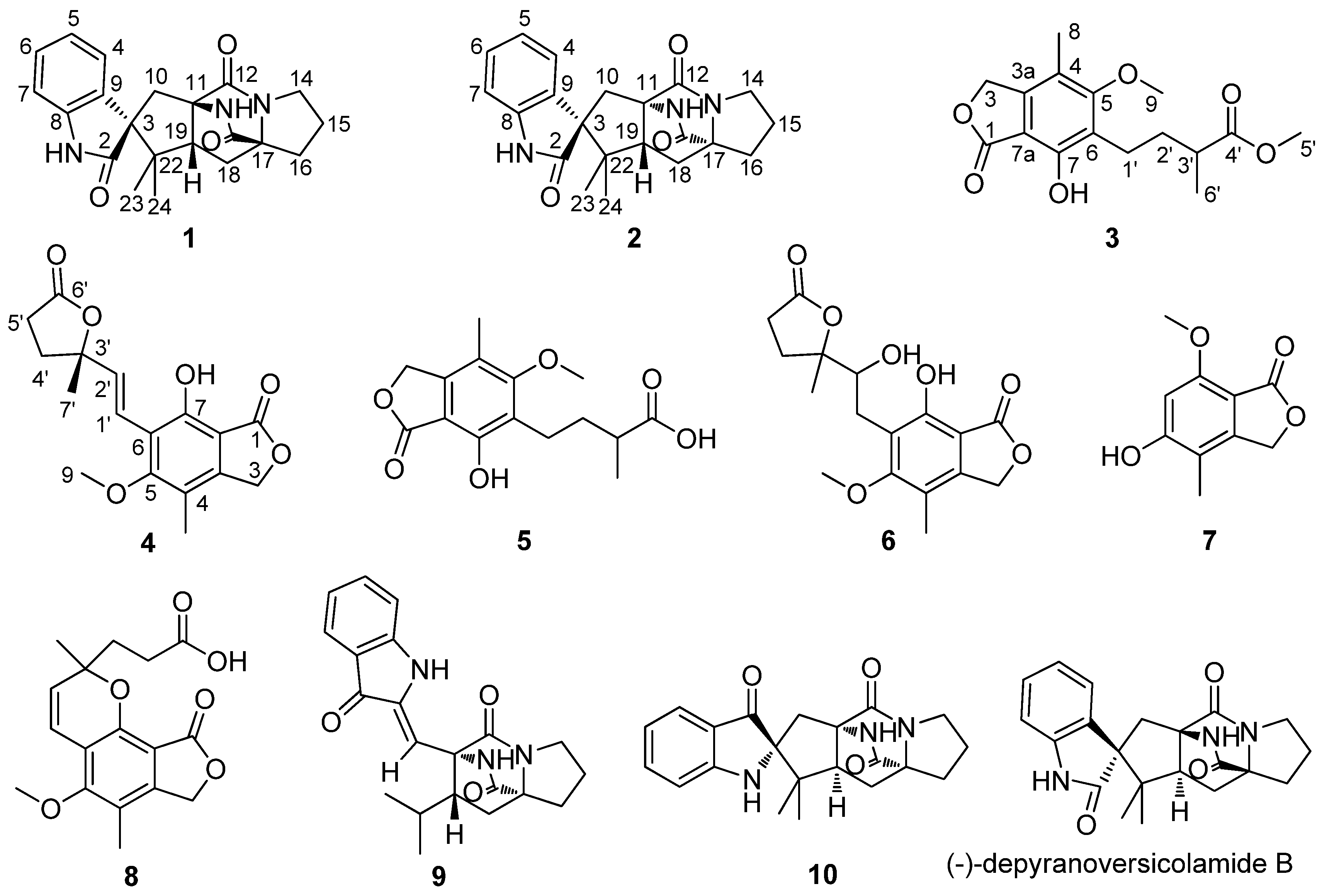
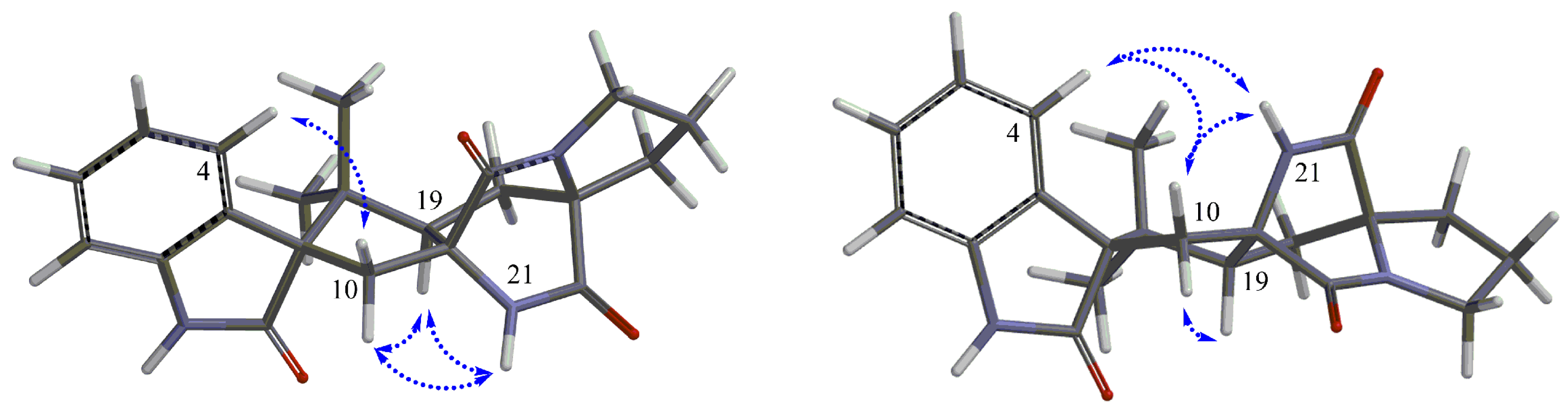
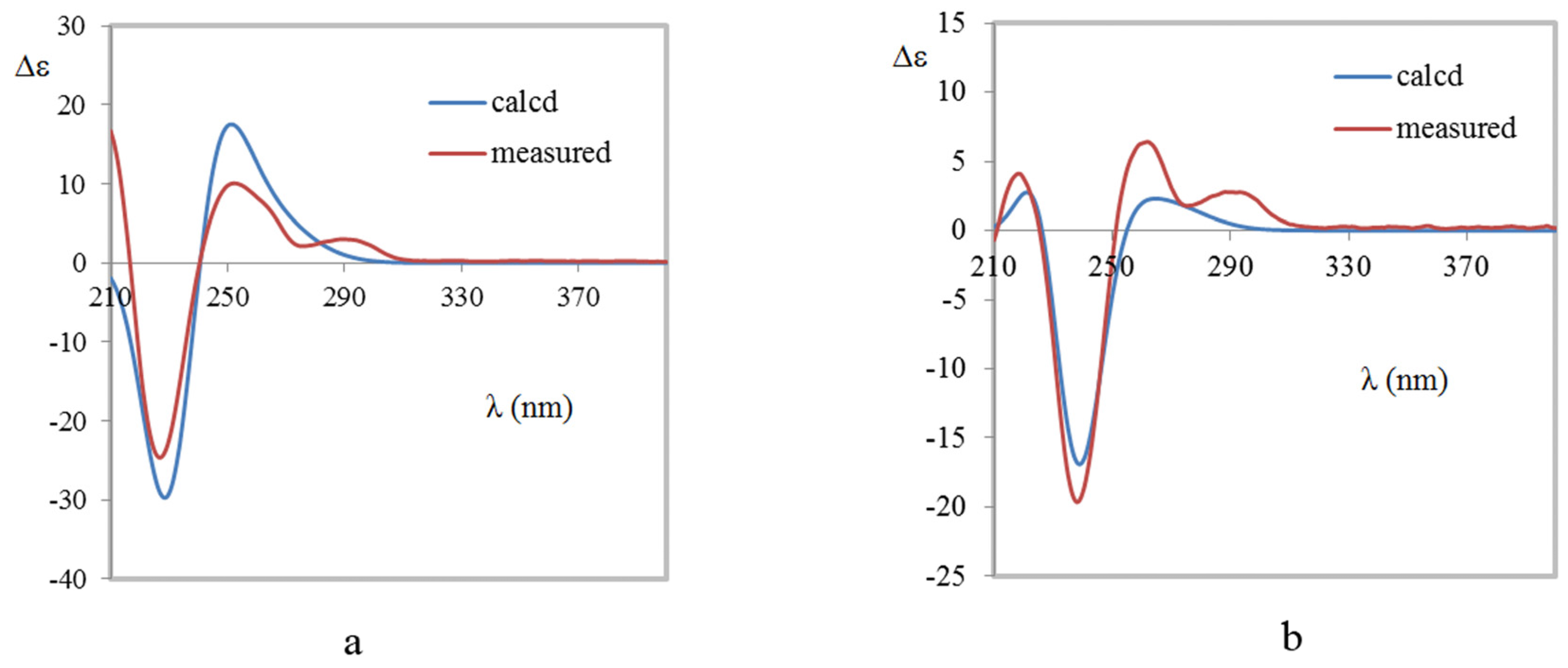
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedure
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Purification
3.5. Computational Methods
3.6. Cytotoxicity
3.7. Larval Settlement Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Z.; Li, S.; Yang, T.; Zhang, Q.; Ma, L.; Tian, X.; Zhang, H.; Huang, C.; Zhang, S.; et al. Spiroindimicins A–D: New bisindole alkaloids from a deep-sea-derived Actinomycete. Org. Lett. 2012, 14, 3364–3367. [Google Scholar] [CrossRef] [PubMed]
- Birch, A.J.; Wright, J.J. The brevianamides: A new class of fungal alkaloid. J. Chem. Soc. D. 1969. [Google Scholar] [CrossRef]
- Paterson, R.R.M.; Simmonds, M.J.S.; Kemmelmeier, C.; Blaney, W.M. Effects of brevianamide A, its photolysis product brevianamide D, and ochratoxin A from two Penicillium strains on the insect pests Spodoptera frugiperda and Heliothis virescens. Mycol. Res. 1990, 94, 538–542. [Google Scholar] [CrossRef]
- Song, F.; Liu, X.; Guo, H.; Ren, B.; Chen, C.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M.; et al. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.F.; Xiao, T.; Zhang, D.; Deng, L.F.; Wang, Y.; Qin, Y. Total synthesis of (-)-depyrannoversicolamide B. Chem. Commun. 2015, 51, 16143–16146. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Glinka, T.; Kwast, E.; Coffman, H.; Stille, J.K. Asymmetric, stereocontrolled total synthesis of (-)-brevianamide B. J. Am. Chem. Soc. 1990, 112, 808–821. [Google Scholar] [CrossRef]
- Zhao, L.; May, J.P.; Huang, J.; Perrin, D.M. Stereoselective synthesis of brevianamide E. Org. Lett. 2012, 14, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.F.; Moore, R.H.; Crawley, G.C. Microbial modification of mycophenolic acid. J. Chem. Soc. C. 1970. [Google Scholar] [CrossRef]
- Sintchak, M.D.; Fleming, M.A.; Futer, O.; Raybuck, S.A.; Chambers, S.P.; Caron, P.R.; Murcko, M.A.; Wilson, K.P. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell 1996, 85, 921–930. [Google Scholar] [CrossRef]
- Bentley, R. Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000, 100, 3801–3825. [Google Scholar] [CrossRef] [PubMed]
- Birch, A.J.; Wright, J.J. Biosynthesis. XLII. Structural elucidation and same aspects of the biosynthesis of the brevianamides-A and -E. Tetrahedron 1970, 26, 2329–2344. [Google Scholar] [CrossRef]
- Nugroho, A.E.; Morita, H. Circular dichroism calculation for natural products. J. Nat. Prod. 2014, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.P.; Liang, X.R.; Liu, X.H.; Ji, N.Y. Aspewentins A-C, norditerpenes from a cryptic pathway in an algicolous strain of Aspergillus wentii. J. Nat. Prod. 2014, 77, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.; Jakupovic, J.; Bohlmann, F. A new type of sesquiterpene and acorane derivative from Calea prunifolia. J. Nat. Prod. 1984, 47, 802–808. [Google Scholar] [CrossRef]
- Mohapatra, D.K.; Pramanik, C.; Chorghade, M.S.; Gurjar, M.K. A short and efficient synthetic strategy for the total syntheses of (S)-(+)- and (R)-(−)-plalolide A. Eur. J. Org. Chem. 2007, 5059–5063. [Google Scholar] [CrossRef]
- Habib, E.; Leon, F.; Bauer, J.D.; Hill, R.A.; Carvalho, P.; Cutler, H.G.; Cutler, S.J. Mycophenolic derivatives from Eupenicillium parvum. J. Nat. Prod. 2008, 71, 1915–1918. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.M.M.P.; Ferreira, A.G.; Daolio, C.; Filho, E.R.; Boffo, E.; Souza, A.Q.L.; Sebastianes, F.L.S.; Melo, I.S. Production of 5-hydroxy-7-methoxy-4-methylphthalide in a culture of Penicillium crustosum. An. Acad. Bras. Cienc. 2013, 85, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Nedachi, M.; Maekawa, M.; Murayama, Y.; Miyazawa, M.; Hirai, Y. Synthetic study for two 2H-chromenic acids, 8-chlorocannabiorcichromenic acid and mycochromenic acid. J. Heterocycl. Chem. 2006, 43, 29–41. [Google Scholar] [CrossRef]
- Birch, A.J.; Russell, R.A. Structural elucidations of brevianamides-B, -C, -D and -F. Tetrahedron 1972, 28, 2999–3008. [Google Scholar] [CrossRef]
- Williams, R.M.; Sanz-Cervera, J.F.; Sancenon, F.; Marco, J.A.; Halligan, K.M. Biomimetic Diels-Alder cyclizations for the construction of the brevianamide, paraherquamide, sclerotamide, asperparaline and VM55599 ring systems. Bioorg. Med. Chem. 1998, 6, 1233–1241. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic-behavior. Phys. Rev. A Gen. Phys. 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yang, W.T.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef] [PubMed]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 mode. J. Chem. Phys. 1999, 110, 6158–6169. [Google Scholar] [CrossRef]
- Staroverov, V.N.; Scuseria, G.E.; Tao, J.; Perdew, J.P. Efficient hybrid density functional calculations in solids: Assessment of the Heyd-Scuseria-Ernzerhof screened Coulomb hybrid functional. J. Chem. Phys. 2003, 119, 12129. [Google Scholar] [CrossRef]
- Schäfer, A.; Huber, C.; Ahlrichs, R. Fully optimized contracted gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Wang, J.; Zhang, J.J.; Song, X.; Ku, C.F.; Zou, J.; Li, J.X.; Rong, L.J.; Pan, L.T.; Zhang, H.J. Henrin A: A new anti-HIV ent-kaurane diterpene from Pteris henryi. Int. J. Mol. Sci. 2015, 16, 27978–27987. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.H.; Xu, Y.; Xiong, H.R.; Qian, P.Y.; Zhang, S. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J. Microbiol. Biotechnol. 2009, 25, 399–406. [Google Scholar] [CrossRef]


| No. | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1-NH | - | 10.36 s | - | 10.31 s |
| 2 | 182.8 C | 182.3 C | ||
| 3 | 61.9 C | 62.6 C | ||
| 4 | 126.4 CH | 7.23, d (7.5) | 126.8 CH | 7.43, d (7.5) |
| 5 | 121.5 CH | 6.98, dd (7.5, 7.6) | 121.3 CH | 6.99, dd (7.5, 7.6) |
| 6 | 128.3 CH | 7.20, dd (7.6, 7.6) | 128.6 CH | 7.20, dd (7.6, 7.6) |
| 7 | 109.6 CH | 6.83, d (7.6) | 109.5 CH | 6.81, d (7.6) |
| 8 | 142.8 C | 142.9 C | ||
| 9 | 131.0 C | 130.2 C | ||
| 10 | 33.6 CH2 | 2.20, d (14.2) | 34.1 CH2 | 2.14, d (15.2) |
| 2.86, d (14.2) | 2.83, d (15.2) | |||
| 11 | 66.1 C | 67.6 C | ||
| 12 | 169.8 C | 169.5 C | ||
| 14 | 43.8 CH2 | 3.41, m | 43.7 CH2 | 3.30, m |
| 15 | 24.8 CH2 | 1.80, m | 24.9 CH2 | 1.83, m |
| 1.99, m | 2.00, dd (5.9, 12.1) | |||
| 16 | 29.5 CH2 | 2.50, overlapped | 28.9 CH2 | 1.79, m |
| 2.47, dd (6.4, 12.1) | ||||
| 17 | 68.5 C | 69.1 C | ||
| 18 | 29.9 CH2 | 1.78, dd (8.2, 12.8) | 28.4 CH2 | 1.79, m |
| 1.93, dd (10.4, 12.9) | 2.47, dd (6.4, 12.1) | |||
| 19 | 55.9 CH | 3.23, dd (8.3, 10.1) | 50.5 CH | 3.18, dd (5.0, 10.0) |
| 20 | 173.5 C | 173.0 C | ||
| 21-NH | - | 9.13, s | - | 8.81, s |
| 22 | 45.6 C | 47.3 C | ||
| 23 | 20.2 CH3 | 0.74, s | 20.9 CH3 | 1.00, s |
| 24 | 23.7 CH3 | 0.72, s | 23.5 CH3 | 0.69, s |
| No. | 3 a | 4 b | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 172.9 C | 170.6 C | ||
| 3 | 70.0 CH2 | 5.18, s | 69.2 CH2 | 5.30, s |
| 3a | 144.1 C | 147.5 C | ||
| 4 | 116.7 C | 117.0 C | ||
| 5 | 163.8 C | 162.8 C | ||
| 6 | 122.5 C | 117.9 C | ||
| 7 | 153.7 C | 153.5 C | ||
| 7a | 106.3 C | 107.8 C | ||
| 8 | 11.6 CH3 | 2.13, s | 11.3 CH3 | 2.09, s |
| 9 | 61.1 CH3 | 3.77, s | 60.6 CH3 | 3.67, s |
| 1′ | 21.3 CH2 | 2.66, m | 118.2 CH | 6.63, d (16.5) |
| 2′ | 33.1 CH2 | 1.90, m | 137.4 CH | 6.76, d (16.1) |
| 1.67, m | ||||
| 3′ | 39.4 CH | 2.50, dq (6.9, 6.9) | 85.8 C | |
| 4′ | 177.0 C | 34.0 CH2 | 2.24, m | |
| 2.19, m | ||||
| 5′ | 51.6 CH3 | 3.68, s | 28.8 CH2 | 2.64, m |
| 2.56, m | ||||
| 6′ | 17.1 CH3 | 1.21 d (1.2) | 177.0 C | |
| 7′ | 26.4 CH3 | 1.55, s | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhang, X.; Nong, X.; Wang, J.; Qi, S. Brevianamides and Mycophenolic Acid Derivatives from the Deep-Sea-Derived Fungus Penicillium brevicompactum DFFSCS025. Mar. Drugs 2017, 15, 43. https://doi.org/10.3390/md15020043
Xu X, Zhang X, Nong X, Wang J, Qi S. Brevianamides and Mycophenolic Acid Derivatives from the Deep-Sea-Derived Fungus Penicillium brevicompactum DFFSCS025. Marine Drugs. 2017; 15(2):43. https://doi.org/10.3390/md15020043
Chicago/Turabian StyleXu, Xinya, Xiaoyong Zhang, Xuhua Nong, Jie Wang, and Shuhua Qi. 2017. "Brevianamides and Mycophenolic Acid Derivatives from the Deep-Sea-Derived Fungus Penicillium brevicompactum DFFSCS025" Marine Drugs 15, no. 2: 43. https://doi.org/10.3390/md15020043





