Isolation of Petrocidin A, a New Cytotoxic Cyclic Dipeptide from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT348
Abstract
:1. Introduction
2. Results and Discussion
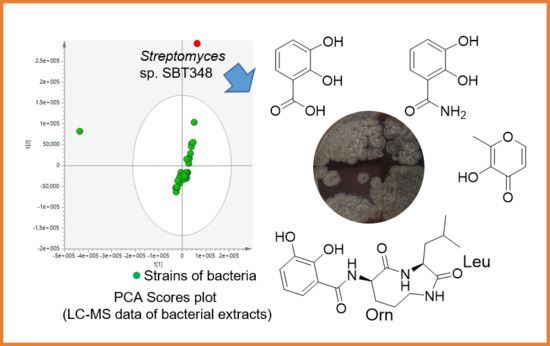
2.1. Isolation and Purification of Strain Streptomyces sp. SBT348
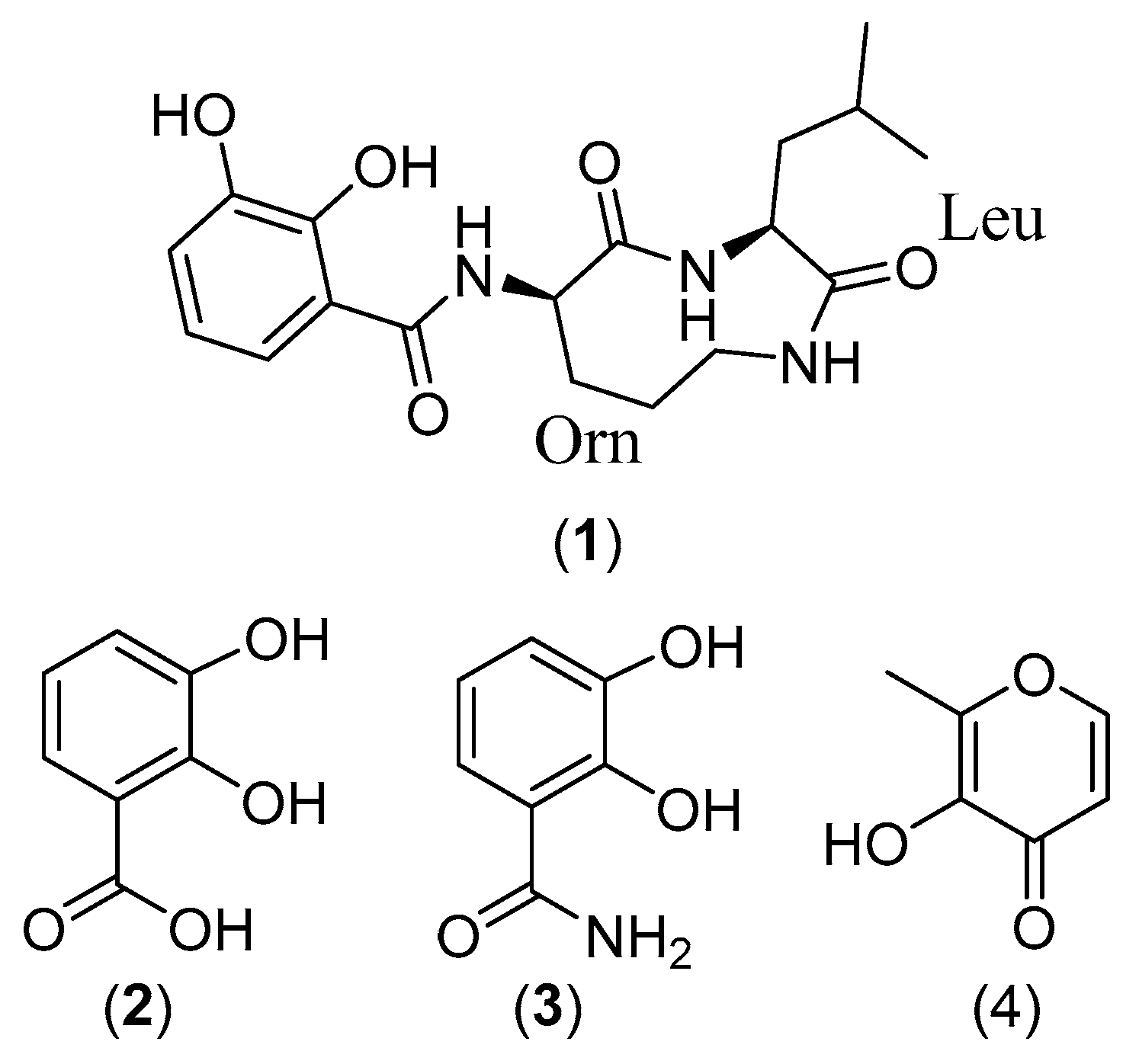
2.2. Structure Elucidation
2.3. Biological Activities of Isolated Compounds
3. Materials and Methods
3.1. Cell Lines, Chemicals, and Biochemical
3.2. Bacteria
3.3. Large-Scale Fermentation, Extraction, and Isolation
3.4. LC-MS Analysis
3.5. Marfey’s Analysis
3.6. MTT ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)) Assay
3.7. Compounds Characterization
3.7.1. Petrocidin A (1)
3.7.2. 2,3-Dihydroxybenzoic Acid (2)
3.7.3. 2,3-Dihydroxybenzamide (3)
3.7.4. Maltol (4)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nouioui, I.; Ruckert, C.; Willemse, J.; van Wezel, G.P.; Klenk, H.P.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Actinoalloteichus fjordicus sp. nov. isolated from marine sponges: Phenotypic, chemotaxonomic and genomic characterisation. Anton Leeuwenhoek 2017, 110, 1705–1717. [Google Scholar] [CrossRef] [PubMed]
- De Menezes, C.B.A.; Afonso, R.S.; de Souza, W.R.; Parma, M.; de Melo, I.S.; Zucchi, T.D.; Fantinatti-Garboggini, F. Williamsia spongiae sp nov., an actinomycete isolated from the marine sponge Amphimedon viridis. Int. J. Syst. Evol. Microbiol. 2017, 67, 1260–1265. [Google Scholar]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Romero, C.; Rode, J.E.; Perez, Y.M.; Franzblau, S.G.; Rodriguez, A.D. Exploring the Sponge Consortium Plakortis symbiotica-Xestospongia deweerdtae as a Potential Source of Antimicrobial Compounds and Probing the Pharmacophore for Antituberculosis Activity of Smenothiazole A by Diverted Total Synthesis. J. Nat. Prod. 2017, 80, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [PubMed]
- Eltamany, E.E.; Abdelmohsen, U.R.; Ibrahim, A.K.; Hassanean, H.A.; Hentschel, U.; Ahmed, S.A. New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 2014, 24, 4939–4942. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Cheng, C.; Viegelmann, C.; Zhang, T.; Grkovic, T.; Ahmed, S.; Quinn, R.J.; Hentschel, U.; Edrada-Ebel, R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs 2014, 12, 1220–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viegelmann, C.; Parker, J.; Ooi, T.; Clements, C.; Abbott, G.; Young, L.; Kennedy, J.; Dobson, A.D.; Edrada-Ebel, R. Isolation and identification of antitrypanosomal and antimycobacterial active steroids from the sponge Haliclona simulans. Mar. Drugs 2014, 12, 2937–2952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabares, P.; Pimentel-Elardo, S.M.; Schirmeister, T.; Hunig, T.; Hentschel, U. Anti-protease and immunomodulatory activities of bacteria associated with Caribbean sponges. Mar. Biotechnol. (N. Y.) 2011, 13, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Reimer, A.; Blohm, A.; Quack, T.; Grevelding, C.G.; Kozjak-Pavlovic, V.; Rudel, T.; Hentschel, U.; Abdelmohsen, U.R. Inhibitory activities of the marine streptomycete-derived compound SF2446A2 against Chlamydia trachomatis and Schistosoma mansoni. J. Antibiot. 2015, 68, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Grkovic, T.; Abdelmohsen, U.R.; Othman, E.M.; Stopper, H.; Edrada-Ebel, R.; Hentschel, U.; Quinn, R.J. Two new antioxidant actinosporin analogues from the calcium alginate beads culture of sponge-associated Actinokineospora sp. strain EG49. Bioorg. Med. Chem. Lett. 2014, 24, 5089–5092. [Google Scholar] [CrossRef] [PubMed]
- Yi-Lei, N.; Yun-Dan, W.; Chuan-Xi, W.; Ru, L.; Yang, X.; Dong-Sheng, F.; Hong, J.; Yun-Yang, L. Compounds from marine-derived Verrucosispora sp. FIM06054 and their potential antitumour activities. Nat. Prod. Res. 2014, 28, 2134–2139. [Google Scholar] [CrossRef] [PubMed]
- Simmons, L.; Kaufmann, K.; Garcia, R.; Schwar, G.; Huch, V.; Muller, R. Bendigoles D-F, bioactive sterols from the marine sponge-derived Actinomadura sp. SBMs009. Bioorg. Med. Chem. 2011, 19, 6570–6575. [Google Scholar] [CrossRef] [PubMed]
- Inui, T.; Wang, Y.; Pro, S.M.; Franzblau, S.G.; Pauli, G.F. Unbiased evaluation of bioactive secondary metabolites in complex matrices. Fitoterapia 2012, 83, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, D.; Muller, R. Secondary metabolomics: The impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 2014, 31, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.; Beale, M.; Fiehn, O.; Hardy, N.; Sumner, L.; Bino, R. Plant Metabolomics: The missing link in functional genomics strategies. Plant Cell 2002, 14, 1437–1440. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; MacIntyre, L.; Abdelmohsen, U.R.; Horn, H.; Polymenakou, P.N.; Edrada-Ebel, R.; Hentschel, U. Biodiversity, anti-trypanosomal activity screening, and metabolomic profiling of actinomycetes isolated from Mediterranean sponges. PLoS ONE 2015, 10, e0138528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbin, J.L.; Bulen, W.A. The isolation and identification of 2,3-dihydroxybenzoic acid and 2-N,6-N-di-92,3-dihydroxybenzoyl)-l-lysine formed by iron-deficient Azotobacter vinelandii. Biochemistry 1969, 8, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Hirota, A. New potent DPPH radical scavengers from a marine-derived actinomycete strain USF-TC31. Biosci. Biotechnol. Biochem. 2009, 73, 2731–2734. [Google Scholar] [CrossRef] [PubMed]
- Samejo, M.Q.; Ndukwe, G.I.; Burdi, D.K.; Bhanger, M.I.; Khan, K.M. Isolation and crystal structure of maltol from Abies pindrow. J. Med. Plants Res. 2009, 3, 55–60. [Google Scholar]
- Feistner, G.J.; Beaman, B.L. Characterization of 2,3-dihydroxybenzoic acid from Nocardia asteroides GUH-2. J. Bacteriol. 1987, 169, 3982–3987. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, N.; Kimura, R.; Murata, T. Isolation of maltol and pharmacological action of maltol and ethyl maltol. Chem. Pharm. Bull. 1974, 22, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Pollack, J.R.; Ames, B.N.; Neilands, J.B. Iron transport in Salmonella typhimurium: Mutants blocked in the biosynthesis of enterobactin. J. Bacteriol. 1970, 104, 635–639. [Google Scholar] [PubMed]
- Chen, Q.; Actis, L.A.; Tolmasky, M.E.; Crosa, J.H. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J. Bacteriol. 1994, 176, 4226–4234. [Google Scholar] [CrossRef] [PubMed]
- Soengas, R.G.; Anta, C.; Espada, A.; Paz, V.; Ares, I.R.; Balado, M.; Rodríguez, J.; Lemos, M.L.; Jiménez, C. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett. 2006, 47, 7113–7116. [Google Scholar] [CrossRef]
- Balado, M.; Osorio, C.R.; Lemos, M.L. A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 2006, 152, 3517–3528. [Google Scholar] [CrossRef] [PubMed]
- Ushanova, V.M.; Ziganshin, A.V.; Repyakh, S.M. Isolation of maltol from Siberian fir bark by the carbon dioxide method. Chem. Nat. Comp. 1998, 34, 104–105. [Google Scholar] [CrossRef]
- Dean, F.M. Naturally occuring oxygen ring compounds. Chem. Abstr. 1963, 59, 640–649. [Google Scholar]
- Maskey, R.P.; Shaaban, M.; Grun-Wollny, I.; Laatsch, H. Quinazolin-4-one derivatives from Streptomyces isolates. J. Nat. Prod. 2004, 67, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yin, S.; Nie, D.; Xie, S.; Ma, L.; Wang, X.; Wu, Y.; Xiao, J. MK886 inhibits the proliferation of HL-60 leukemia cells by suppressing the expression of mPGES-1 and reducing prostaglandin E2 synthesis. Int. J. Hematol. 2011, 94, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Elander, N.; Zhou, J.; Ungerback, J.; Dimberg, J.; Soderkvist, P. Association between adenomatosis polyposis coli functional status and microsomal prostaglandin E synthase-1 expression in colorectal cancer. Mol. Carcinog. 2009, 48, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Lauro, G.; Tortorella, P.; Bertamino, A.; Ostacolo, C.; Koeberle, A.; Fischer, K.; Bruno, I.; Terracciano, S.; Gomez-Monterrey, I.M.; Tauro, M.; et al. Structure-based design of microsomal prostaglandin E2 Synthase-1 (mPGES-1) inhibitors using a virtual fragment growing optimization scheme. ChemMedChem 2016, 11, 612–619. [Google Scholar] [CrossRef] [PubMed]
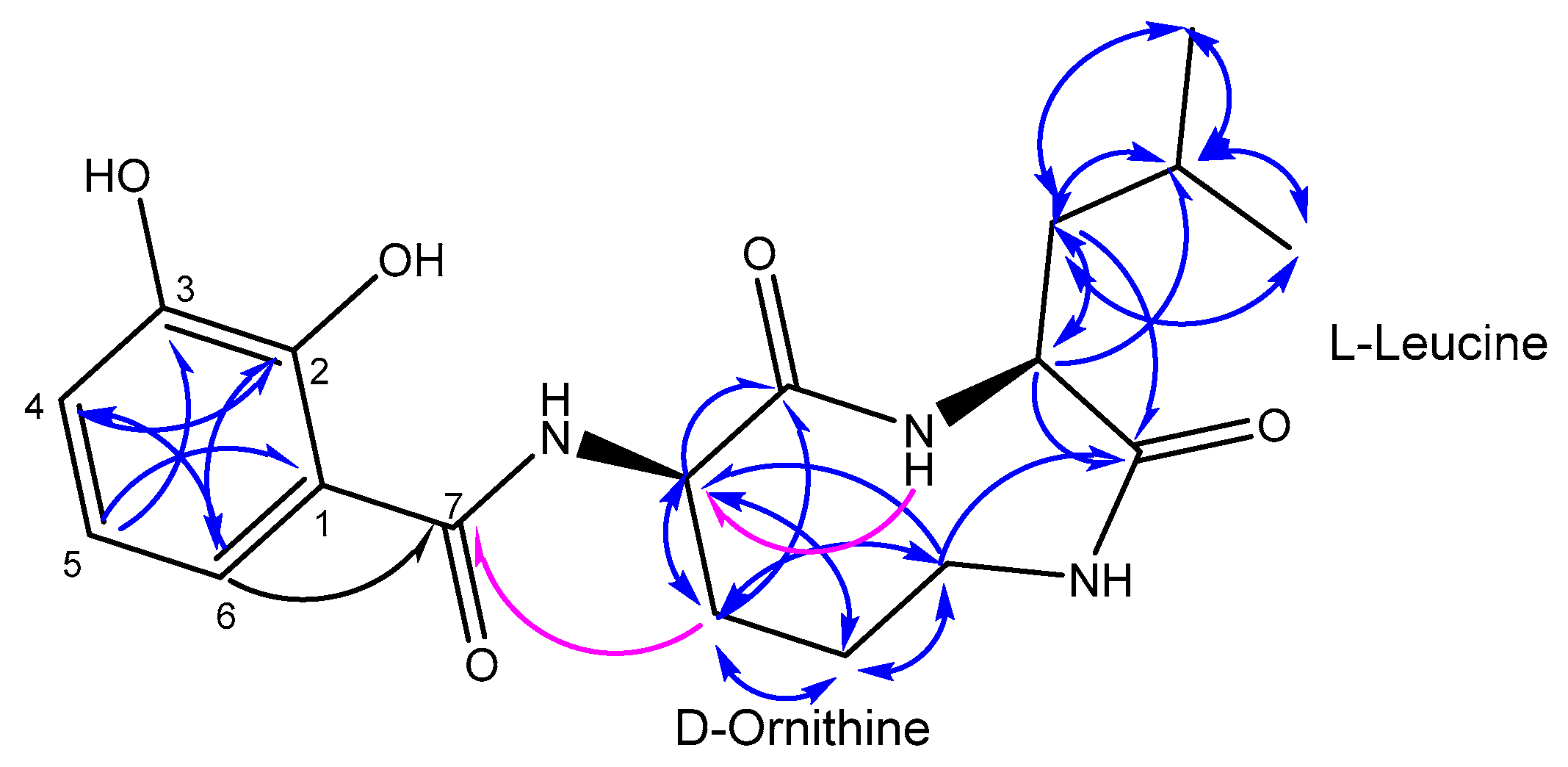
| Position | δC, Type | δH (J, Hz) | COSY | HMBC a | HMBC b |
|---|---|---|---|---|---|
| 1 | 114.7, C | - | - | - | - |
| 2 | 151.7, C | - | - | - | - |
| 3 | 147.0, C | - | - | - | - |
| 4 | 121.3, CH | 6.98 (1H, d, J = 7.8 Hz) | 6.71 | 151.7, 147.0, 121.9 | - |
| 5 | 119.6, CH | 6.71 (1H, t, J = 7.8 Hz) | 6.98, 7.34 | 147.0, 114.7, 121.3 | - |
| 6 | 121.9, CH | 7.34 (1H, d, J = 7.8 Hz) | 6.71 | 174.2, 151.7, 121.3 | - |
| 7 | 174.2, C | - | - | - | - |
| Orn | - | - | - | - | - |
| CO | 172.8, C | - | - | - | - |
| α | 60.3, CH | 4.26 (1H, t, J = 8.5 Hz) | 2.30, 2.01 | 172.8, 29.1, 23.7 | - |
| β | 29.1, CH2 | 2.30 (1H, m), 2.01 (1H, m) | 2.02, 1.94 | 46.4, 23.7, 60.3, 172.8 | 171.9 (C-7) |
| γ | 23.7, CH2 | 2.02 (1H, m), 1.94 (1H, m) | 2.30, 2.01, 2.02, 1.94 | 29.1, 60.3, 46.4 | - |
| δ | 46.4, CH2 | 3.51 (2H, t, J = 4.7 Hz) | 2.02, 1.94 | 168.9, 29.1, 23.7 | - |
| Leu | - | - | - | - | - |
| NH | - | 8.02 (1H, s, DMSO-d6) | - | - | 37.8 (Leu β-C), 52.6 (Leu α-C), 58.5 (Orn α-C), 166.6 (Leu CO) |
| CO | 168.9, C | - | - | - | - |
| α | 54.6, CH | 4.13 (1H, t, J = 5.6 Hz) | 1.91, 1.52 | 168.9, 39.4, 25.8 | - |
| β | 39.4, CH2 | 1.52 (1H, m), 1.91 (1H, m) | 1.89, 4.13 | 168.9, 54.6, 25.8, 23.2, 22.2 | - |
| γ | 25.8, CH | 1.89 (1H, m) | 0.96, 0.95, 1.52, 1.91 | 39.4 | - |
| δ | 22.2, 23.2, CH3 | 0.96 (3H, d, J = 6.5 Hz) | 1.89 | 25.8, 39.4 | - |
| 0.95 (3H, d, J = 3.1, 6.2 Hz) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; Othman, E.M.; Stopper, H.; Edrada-Ebel, R.; Hentschel, U.; Abdelmohsen, U.R. Isolation of Petrocidin A, a New Cytotoxic Cyclic Dipeptide from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT348. Mar. Drugs 2017, 15, 383. https://doi.org/10.3390/md15120383
Cheng C, Othman EM, Stopper H, Edrada-Ebel R, Hentschel U, Abdelmohsen UR. Isolation of Petrocidin A, a New Cytotoxic Cyclic Dipeptide from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT348. Marine Drugs. 2017; 15(12):383. https://doi.org/10.3390/md15120383
Chicago/Turabian StyleCheng, Cheng, Eman M. Othman, Helga Stopper, RuAngelie Edrada-Ebel, Ute Hentschel, and Usama Ramadan Abdelmohsen. 2017. "Isolation of Petrocidin A, a New Cytotoxic Cyclic Dipeptide from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT348" Marine Drugs 15, no. 12: 383. https://doi.org/10.3390/md15120383