Natural Products from Microalgae with Potential against Alzheimer’s Disease: Sulfolipids Are Potent Glutaminyl Cyclase Inhibitors
Abstract
:1. Introduction
2. Results
3. Discussion
- Metal binding group (MBG)
- Flexible linker with minimum length (propyl-linker)
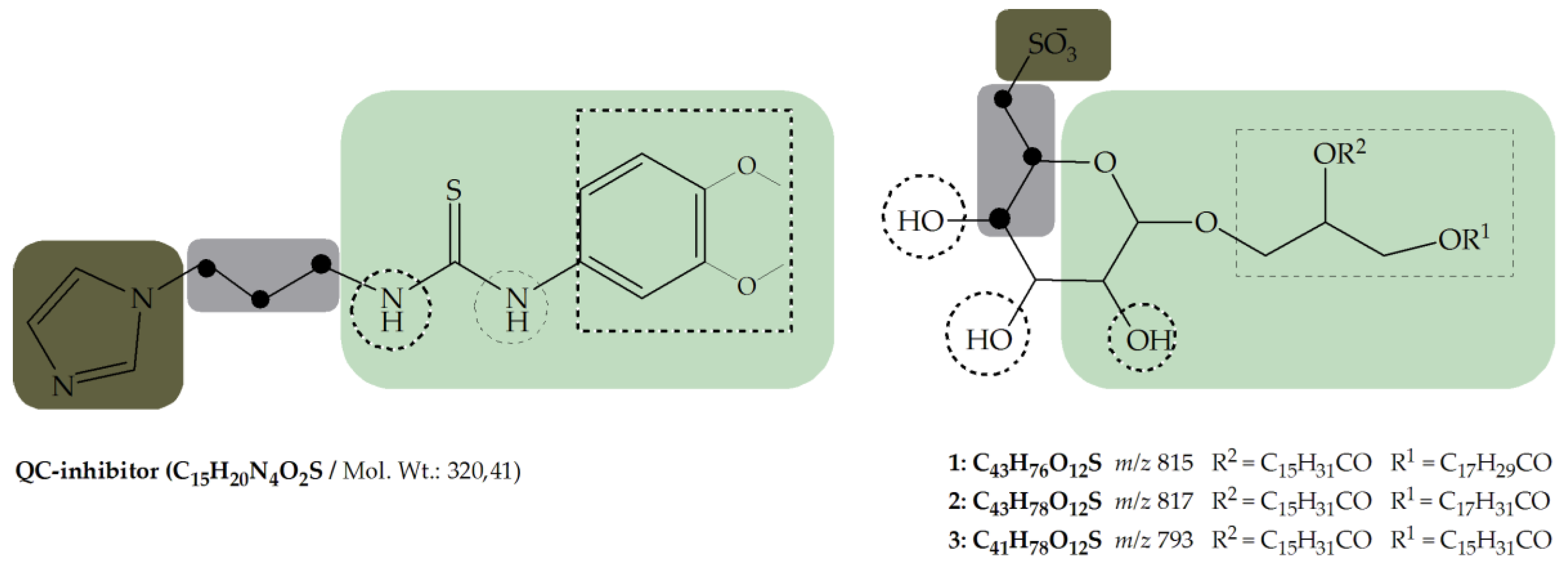
- Core structure (scaffold) decorated with functional groups at certain positions e.g., 3,4-dimethoxyphenylthiourea, which additionally can form hydrogen bonds and lipophilic interactions within the enzyme pocket.
4. Materials and Methods
4.1. Cultivation of the Microalgae
4.2. Preparation of Crude Extracts
4.3. Sample Preparation
4.4. Glutaminyl Cyclase (QC) Inhibition Assay
4.5. Mass Spectrometry
4.5.1. UPLC/ESI-MS
4.5.2. ESI-FTICR MS
4.6. MS Data Processing and Activity Correlation Analysis (AcorA)
4.7. Isolation of Sulfolipids
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AcorA | Activity-correlation Analysis |
| AChE | acetylcholine esterase |
| APP | amyloid precursor protein |
| AD | Alzheimer’s disease |
| BACE-1 | Beta-secretase 1; beta-site amyloid precursor protein cleaving enzyme 1 |
| calcd. | calculated |
| CH3COOH | acetic acid |
| ESI-FTICR MS | electrospray ionization Fourier transform ion cyclotron mass spectrometry |
| Gln-AMC | N-glutaminyl-7-amino-4-methyl-coumarin |
| GP | exponential growth phase |
| Isotope p. | isotope peak |
| MBG | metal binding group |
| NaOAc | sodium acetat |
| NH4OAc | ammonium acetate |
| pGAP | pyroglutaminyl aminopeptidase |
| pGlu | pyroglutamyl |
| rs | Spearman’s rank correlation coefficient |
| RT | retention time |
| SP | stationary growth phase |
| QC | glutaminyl cyclase |
| UPLC-MS | Ultra performance liquid chromatography mass spectrometry |
References
- Busby, W.H.; Quackenbush, G.E.; Humm, J.; Youngblood, W.W.; Kizers, J.S. An Enzyme (s) That Converts Glutaminyl-peptides into pyroglutamyl-petides. J. Biol. Chem. 1987, 262, 8532–8536. [Google Scholar] [PubMed]
- Fischer, W.H.; Spiess, J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc. Natl. Acad. Sci. USA 1987, 84, 3628–3632. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Niestroj, A.J.; Rahfeld, J.-U.; Hoffmann, T.; Wermann, M.; Zunkel, K.; Wasternack, C.; Demuth, H.-U. Identification of human glutaminyl cyclase as a metalloenzyme. Potent inhibition by imidazole derivatives and heterocyclic chelators. J. Biol. Chem. 2003, 278, 49773–49779. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.N.; Podell, D.N. Pyroglutamic acid-Non-metabolic formation, function in proteins and peptides, and characteristics of the enzymes effecting its removal. Mol. Cell. Biochem. 1981, 38, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Folkers, K.; Chang, J.K.; Currie, B.L.; Bowers, C.Y.; Weil, A.; Schally, A.V. Synthesis and relationship of l-glutaminyl-l-histidyl-l-prolinamide to the thyrotropin releasing hormone. Biochem. Biophys. Res. Commun. 1970, 39, 110–113. [Google Scholar] [CrossRef]
- Messer, M.; Ottesen, M. Isolation and properties of glutamine cyclotransferase of dried papaya latex. Biochim. Biophys. Acta (BBA)-Spec. Sect. Enzymol. Subj. 1964, 92, 409–411. [Google Scholar] [CrossRef]
- El Moussaoui, A.; Nijs, M.; Paul, C.; Wintjens, R.; Vincentelli, J.; Azarkan, M.; Looze, Y. Revisiting the enzymes stored in the laticifers of Carica papaya in the context of their possible participation in the plant defence mechanism. Cell. Mol. Life Sci. 2001, 58, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Böckers, T.M.; Kreutz, M.R.; Pohl, T. Glutaminyl-Cyclase Expression in the Bovine/Porcine Hypothalamus and Pituitary. J. Neuroendocrinol. 1995, 7, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Dahl, S.W.; Slaughter, C.; Lauritzen, C.; Bateman, R.C.; Connerton, I.; Pedersen, J. Carica papaya glutamine cyclotransferase belongs to a novel plant enzyme subfamily: Cloning and characterization of the recombinant enzyme. Protein Expr. Purif. 2000, 20, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Pohl, T.; Zimmer, M.; Mugele, K.; Spiess, J. Primary structure and functional expression of a glutaminyl cyclase. Proc. Natl. Acad. Sci. USA 1991, 88, 10059–10063. [Google Scholar] [CrossRef] [PubMed]
- Song, I.; Chuang, C.Z.; Bateman, R.C.J. Molecular cloning, sequence analysis and expression of human pituitary glutmaniyl cyclase. J. Mol. Endocrinol. 1994, 13, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Hoffmann, T.; Rosche, F.; Wasternack, C.; Demuth, H.; Manhart, S. Heterologous Expression and Characterization of Human Glutaminyl Cyclase: Evidence for a Disulfide Bond with Importance for Catalytic Activity. Biochemistry 2002, 41, 10849–10857. [Google Scholar] [CrossRef] [PubMed]
- Batliwalla, F.M.; Baechler, E.C.; Xiao, X.; Li, W.; Balasubramanian, S.; Khalili, H.; Damle, A.; Ortmann, W.; Perrone, A.; Kantor, B.; et al. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun. 2005, 6, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Ezura, Y.; Kajita, M.; Ishida, R.; Yoshida, S.; Yoshida, H.; Suzuki, T.; Hosoi, T.; Inoue, S.; Shiraki, M.; Orimo, H.; et al. Association of multiple nucleotide variations in the pituitary glutaminyl cyclase gene (QPCT) with low radial BMD in adult women. J. Bone Miner. Res. 2004, 19, 1296–1301. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Hoffmann, T.; Manhart, S.; Hoffmann, M.; Demuth, H.-U. Glutaminyl cyclases unfold glutamyl cyclase activity under mild acid conditions. FEBS Lett. 2004, 563, 191–196. [Google Scholar] [CrossRef]
- Cynis, H.; Schilling, S.; Bodnár, M.; Hoffmann, T.; Heiser, U.; Saido, T.C.; Demuth, H.-U. Inhibition of glutaminyl cyclase alters pyroglutamate formation in mammalian cells. Biochim. Biophys. Acta 2006, 1764, 1618–1625. [Google Scholar] [CrossRef] [PubMed]
- Cynis, H.; Scheel, E.; Saido, T.C.; Schilling, S.; Demuth, H.U. Amyloidogenic processing of amyloid precursor protein: Evidence of a pivotal role of glutaminyl cyclase in generation of pyroglutamate-modified amyloid-β. Biochemistry 2008, 47, 7405–7413. [Google Scholar] [CrossRef] [PubMed]
- Saido, T.C. Alzheimer’s disease as proteolytic disorders: Anabolism and catabolism of β-amyloid. Neurobiol. Aging 1998, 19, 69–75. [Google Scholar] [CrossRef]
- Russo, C.; Violani, E.; Salis, S.; Venezia, V.; Dolcini, V.; Damonte, G.; Benatti, U.; D’Arrigo, C.; Patrone, E.; Carlo, P.; et al. Pyroglutamate-modified amyloid ß-peptides—AβN3(pE)—Strongly affect cultured neuron and astrocyte survival. J. Neurochem. 2002, 82, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Zeitschel, U.; Hoffmann, T.; Heiser, U.; Francke, M.; Kehlen, A.; Holzer, M.; Hutter-Paier, B.; Prokesch, M.; Windisch, M.; et al. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer’s disease-like pathology. Nat. Med. 2008, 14, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Appl, T.; Hoffmann, T.; Cynis, H.; Schulz, K.; Jagla, W.; Friedrich, D.; Wermann, M.; Buchholz, M.; Heiser, U.; et al. Inhibition of glutaminyl cyclase prevents pGlu-Abeta formation after intracortical/hippocampal microinjection in vivo/in situ. J. Neurochem. 2008, 106, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Jawhar, S.; Wirths, O.; Schilling, S.; Graubner, S.; Demuth, H.-U.; Bayer, T. Overexpression of glutaminyl cyclase, the enzyme responsible for pyroglutamate Aβ formation, induces behavioral deficits, and glutaminyl cyclase knock-out rescues the behavioral phenotype in 5XFAD mice. J. Biol. Chem. 2011, 286, 4454–4460. [Google Scholar] [CrossRef] [PubMed]
- Demuth, H.-U.; Schilling, S.; Roßner, S.; Morawski, M.; Hartlage-Rübsamen, M.; Lues, I.; Glund, K. Toxic Pglu-Abeta Is Enhanced and Glutaminyl Cyclase (Qc) Up-Regulated Early in Alzheimer’s Disease (Ad): Inhibitors of Qc Blocking Pglu-Abeta Formation Are in Clinical Development. Alzheimer’s Dement. 2014, 10, P149. [Google Scholar] [CrossRef]
- Kreuzberger, M. Neuronale Verteilung des Enzyms Glutaminylzyklase im Kortex und der hippocampalen Formation des humanen Gehirns. Ph.D. Thesis, Universität Leipzig, Leipzig, Germany, January 2015. [Google Scholar]
- Höfling, C.; Indrischek, H.; Höpcke, T.; Waniek, A.; Cynis, H.; Koch, B.; Schilling, S.; Morawski, M.; Demuth, H.-U.; Roßner, S.; et al. Mouse strain and brain region-specific expression of the glutaminyl cyclases QC and isoQC. Int. J. Dev. Neurosci. 2014, 36, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Wessjohann, L.A. Reverse metabolomics—Metabolomics in drug discovery: Connecting metabolomic profiles with phylogenetic, medicinal and flavoring properties. Metab. Syst. Biol. 2014, 4, 70. [Google Scholar]
- Degenhardt, A.; Wittlake, R.; Steilwind, S.; Liebig, M.; Runge, C.; Hilmer, J.M.; Krammer, G.; Gohr, A.; Wessjohann, L. Quantification of important flavour compounds in beef stocks and correlation to sensory results by “Reverse Metabolomics”. In Flavour Science; Ferreira, V., Ed.; Elsevier: Amsterdam, The Neatherland, 2013; pp. 15–19. [Google Scholar]
- Krause-Hielscher, S.; Demuth, H.; Wessjohann, L.; Arnold, N.; Griehl, C. Microalgae as a source for potential anti-Alzheimer’s disease directed compounds—Screening for glutaminyl cyclase (QC) inhibiting metabolites. Int. J. Pharm. Biol. Sci. 2015, 5, 164–170. [Google Scholar]
- Zianni, R.; Bianco, G.; Lelario, F.; Losito, I.; Palmisano, F.; Cataldi, T.R.I. Fatty acid neutral losses observed in tandem mass spectrometry with collision-induced dissociation allows regiochemical assignment of sulfoquinovosyl-diacylglycerols. J. Mass Spectrom. 2013, 48, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.A.; Daniel, H.; Wiser, R. A Sulfolipid in Plants. Biochemistry 1959, 45, 1582–1587. [Google Scholar] [CrossRef]
- Lepage, M.; Daniel, H.; Benson, A.A. The Plant Sulfolipid.—Isolation and Properties of Sulfoglycosyl Glycerol. J. Am. Chem. Soc. 1961, 3735, 1958–1960. [Google Scholar]
- Miyano, M.; Benson, A.A. The Plant Sulfolipid. VI. Configuration of the Glycerol Moiety. J. Am. Chem. Soc. 1962, 84, 57–59. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sahara, H.; Fujita, T.; Shimozawa, K.; Takenouchi, M.; Torigoe, T.; Hanashami, S.; Yamazaki, T.; Takahashi, H.; Sugawara, F.; et al. An immunosuppressive effect by synthetic sulfonolipids deduced from sulfonoquinovosyldiacylglycerols of sea urchin. Transplantation 2002, 74, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, D.; Yan, X.; Chen, J.; Zhou, C. Global characterization of the photosynthetic glycerolipids from a marine diatom Stephanodiscus sp. by ultra performance liquid chromatography coupled with electrospray ionization-quadrupole-time of flight mass spectrometry. Anal. Chim. Acta 2010, 663, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Naumann, I.; Darsow, K.H.; Walter, C.; Lange, H.A.; Buchholz, R. Identification of sulfoglycolipids from the alga Porphyridium purpureum by matrix-assisted laser desorption/ionisation quadrupole ion trap time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3185–3192. [Google Scholar] [CrossRef] [PubMed]
- Naumann, I. Sulfoquinovosyldiacylglyceride-Antiviral Aktive Substanzen. Ph.D. Thesis, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, March 2009. [Google Scholar]
- Naumann, I.; Klein, B.; Bartel, C.; Darsow, S.J.; Buchholz, R.; Lange, H.A. Identification of sulfoquinovosyldiacyglycerides from Phaeodactylum tricornutum by matrix-assisted laser desorption/ionization QTrap time-of-flight hybrid mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 2517–2523. [Google Scholar] [CrossRef] [PubMed]
- Keusgen, M.; Curtis, J.M.; Thibault, P.; Walter, J.A.; Windust, A.; Ayer, S.W. Sulfoquinovosyl Diacylglycerols from the Alga Heterosigma carterae. Lipids 1997, 32, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Makewicz, A.; Gribi, C.; Eichenberger, W. Lipids of Ectocarpus fasciculatus (Phaeophyceae). Incorporation of Oleate and the Role of TAG and MGDG in Lipid Metabolism. Plant Cell Physiol. 1997, 38, 952–960. [Google Scholar] [CrossRef]
- Ohta, K.; Mizushina, Y.; Hirata, N.; Takemura, M.; Sugawara, F.; Matsukage, A.; Sakaguchi, K. Sulfoquinovosyldiacylglycerol, KM043, a new potent inhibitor of eukaryotic DNA polymerases and HIV-reverse transcriptase type 1 from a marine red alga, Gigartina tenella. Chem. Pharm. Bull. 1998, 46, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.-L.; Shen, W.-Z.; Rui, W.; Ma, X.-J.; Cen, Y.-Z. Antiviral activity of a sulfoquinovosyldiacylglycerol (SQDG) compound isolated from the green alga Caulerpa racemosa. Bot. Mar. 2007, 50, 185–190. [Google Scholar] [CrossRef]
- Gustafson, K.R.; John, H.; Ii, C.; Fuller, R.W.; Weislow, O.S.; Rebecca, F.; Snader, K.M.; Patterson, G.M.L.; Boyd, M.R. AIDS-Antiviral Sulfolipids From Cyanobacteria (Blue-Green Algae). J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef] [PubMed]
- Reshef, V.; Mizrachi, E.; Maretzki, T.; Silberstein, C.; Loya, S.; Hizi, A.; Carmeli, S. New acylated sulfoglycolipids and digalactolipids and related known glycolipids from cyanobacteria with a potential to inhibit the reverse transcriptase of HIV-1. J. Nat. Prod. 1997, 60, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.A.; Huang, Z.H.; Benning, C. Comparison of sulfoquinovosyl diacylglycerol from spinach and the purple bacterium Rhodobacter spaeroides by fast atom bombardment tandem mass spectrometry. Lipids 1992, 27, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Heinz, E.; Schmidt, H.; Hoch, M.; Jung, K.H.; Binder, H.; Schmidt, R.R. Synthesis of different nucleoside 5′-diphospho-sulfoquinovoses and their use for studies on sulfolipid biosynthesis in chloroplasts. Eur. J. Biochem. 1989, 184, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Seifert, U.; Heinz, E. Enzymatic characteristics of UDP-sulfoquinovose: Diacylglycerol sulfoquinovosyl-transferase from chloroplast envelopes. Bot. Acta 1992, 105, 197–205. [Google Scholar] [CrossRef]
- Pugh, C.E.; Roy, B.; Hawkes, T.; Harwood, J.L. A new pathway for the synthesis of the plant sulpholipid, sulphoquinovosyldiacylglycerol. Biochem. J. 1995, 309, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Bandurski, R. The mechanism of “active sulfate” formation. J. Am. Soc. 1956, 78, 6408–6409. [Google Scholar] [CrossRef]
- Robbins, P.; Lipmann, F. The enzymatic sequence in the biosynthesis of active sulfate. J. Am. Chem. Soc. 1956, 78, 6409–6410. [Google Scholar] [CrossRef]
- Mercer, E.I.; Thomas, G. Occurrence of ATP-adenylsulfate 3′-phosphotransferase in the chloroplasts of higher plants. Phytochemistry 1969, 8, 2281–2285. [Google Scholar] [CrossRef]
- Webb, M.S.; Green, B.R. Biochemical and biophysical properties of thylakoid acyl lipids. Biochim. Biophys. Acta-Bioenerg. 1991, 1060, 133–158. [Google Scholar] [CrossRef]
- Benning, C. Biosynthesis and Function of the Sulfolipid Sulfoquinovosyl Diacylglycerol. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Danishefsky, S.J. Synthesis of a Cyanobacterial Sulfolipid: Confirmation of Its Structure, Stereochemistry, and Anti-HIV-1 Activity. J. Am. Chem. Soc. 1992, 51, 659–663. [Google Scholar] [CrossRef]
- Hanashima, S.; Mizushina, Y.; Yamazaki, T.; Ohta, K.; Takahashi, S.; Koshino, H.; Sahara, H.; Sakaguchi, K.; Sugawara, F. Structural determination of sulfoquinovosyldiacylglycerol by chiral syntheses. Tetrahedron Lett. 2000, 41, 4403–4407. [Google Scholar] [CrossRef]
- Hanashima, S.; Mizushina, Y.; Yamazaki, T.; Ohta, K.; Takahashi, S.; Sahara, H.; Sakaguchi, K.; Sugawara, F. Synthesis of sulfoquinovosylacylglycerols, inhibitors of eukaryotic DNA polymerase alpha and beta. Bioorg. Med. Chem. 2001, 9, 367–376. [Google Scholar] [CrossRef]
- Kurihara, H.; Mitani, T.; Kawabata, J.; Hatano, M. Inhibitory Effect on the α-Glucosidase Reaction by Aggregates State of Sulfoquinovosyldiacylglycerol. Biosci. Biotechnol. Biochem. 1997, 61, 536–538. [Google Scholar] [CrossRef]
- Matsumoto, K.; Sakai, H.; Takeuchi, R.; Tsuchiya, K.; Ohta, K.; Sugawara, F.; Abe, M.; Sakaguchi, K. Effective form of sulfoquinovosyldiacyglycerol (SQDG) vesicles for DNA polymerase inhibition. Colloids Surf. B Biointerfaces 2005, 46, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Brahmi, M.M.; Portmann, C.; D’Ambrosio, D.; Woods, T.M.; Banfi, D.; Reichenbach, P.; da Silva, L.; Baudat, E.; Turcatti, G.; Lingner, J.; et al. Telomerase Inhibitors from Cyanobacteria: Isolation and Synthesis of Sulfoquinovosyl Diacylglycerols from Microcystis aeruguinosa PCC 7806. Chemistry 2013, 19, 4596–4601. [Google Scholar] [CrossRef] [PubMed]
- Bergé, J.P.; Debiton, E.; Dumay, J.; Durand, P.; Barthomeuf, C. In vitro anti-inflammatory and anti-proliferative activity of sulfolipids from the red alga Porphyridium cruentum. J. Agric. Food Chem. 2002, 50, 6227–6232. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Rossi, C.; Marcolongo, G.; di Lena, A.; Venzo, A.; Berrie, C.P.; Corda, D. Selective in vivo anti-inflammatory action of the galactolipid monogalactosyldiacylglycerol. Eur. J. Pharmacol. 2005, 524, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, R.; Singh, O.; Pachuau, L.; Malik, S.P.; Paul, M.; Bhadra, K.; Paul, S.; Kumar, G.S.; Mondal, N.B.; Banerjee, S. Identification of a sulfonoquinovosyldiacylglyceride from Azadirachta indica and studies on its cytotoxic activity and DNA binding properties. Bioorg. Med. Chem. Lett. 2010, 20, 6699–6702. [Google Scholar] [CrossRef] [PubMed]
- Murakami, C.; Yamazaki, T.; Hanashima, S.; Takahashi, S.; Ohta, K.; Yoshida, H.; Sugawara, F.; Sakaguchi, K.; Mizushina, Y. Structure-function relationship of synthetic sulfoquinovosyl-acylglycerols as mammalian DNA polymerase inhibitors. Arch. Biochem. Biophys. 2002, 403, 229–236. [Google Scholar] [CrossRef]
- Noguchi, S.; Akiyama, J.; Hada, A.; Inoue, Y.; Araki, A.; Yukino, T.; Hayashi, M.; Tada, M.; Takahata, K. Effect of Sulfoquinovosyldiacylglycerol (SQDG) extracted from Sea alga (Porphyra yezoensis) on Morphological Differentiation and Apoptosis in Neuro2a Neuroblastoma Cells. Jpn. J. Food Chem 2003, 10, 8530. [Google Scholar]
- Ohta, K.; Miura, M.; Sakaguchi, K. Sulfonated Sugar Compounds, Pharmaceutical Compositions Which contain the Same, and Methods of Treating Tumors with the Same. U.S. Patent 7,973,145 B2, 5 July 2011. [Google Scholar]
- Vasänge, M.; Rolfsen, W.; Bohlin, L. Sulpholipid Composition and Methods for Treating Skin Disorders. U.S. Patent 6,124,266 A, 26 September 2000. [Google Scholar]
- Demuth, H.-U.; Cynis, H.; Alexandru, A.; Jagla, W.; Graubner, S.; Schilling, S. Inhibition of Glutaminyl Cyclase: Pharmacology and steps towards clinical development. Alzheimer’s Dement. 2010, 6, S571–S572. [Google Scholar] [CrossRef]
- Jawhar, S.; Wirths, O.; Bayer, T. A Pyroglutamate amyloid-β (Aβ): A hatchet man in Alzheimer disease. J. Biol. Chem. 2011, 286, 38825–38832. [Google Scholar] [CrossRef] [PubMed]
- Lues, I.; Weber, F.; Meyer, A.; Bühring, U.; Hoffmann, T.; Kühn-Wache, K.; Manhart, S.; Heiser, U.; Pokorny, R.; Chiesa, J.; et al. A phase 1 study to evaluate the safety and pharmacokinetics of PQ912, a glutaminyl cyclase inhibitor, in healthy subjects. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2015, 1, 182–195. [Google Scholar] [CrossRef]
- Buchholz, M.; Heiser, U.; Schilling, S.; Niestroj, A.J.; Zunkel, K.; Demuth, H.-U. The first potent inhibitors for human glutaminyl cyclase: Synthesis and structure-activity relationship. J. Med. Chem. 2006, 49, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, M.; Hamann, A.; Aust, S.; Brandt, W.; Böhme, L.; Hoffmann, T.; Schilling, S.; Demuth, H.-U.; Heiser, U. Inhibitors for human glutaminyl cyclase by structure based design and bioisosteric replacement. J. Med. Chem. 2009, 52, 7069–7080. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, M.; Yurewicz, E.; Patel, G. A new fluorogenic substrate for chymotrypsin. Anal. Biochem. 1976, 70, 258–262. [Google Scholar] [CrossRef]
- Schilling, S.; Hoffmann, T.; Wermann, M.; Heiser, U.; Wasternack, C.; Demuth, H.-U. Continuous spectrometric assays for glutaminyl cyclase activity. Anal. Biochem. 2002, 303, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S. Charakterisierung der Humanen Glutaminyl-Cyclase im Vergleich mit dem Analogen Enzym aus Carica papaya. Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany, May 2004. [Google Scholar]

| Algae Extract | Inhibition of QC Enzyme Activity [%] * cextract = 0.2 mg/mL | Algae Extract | Inhibition of QC Enzyme Activity[%] * cextract = 0.2 mg/mL |
|---|---|---|---|
| Sc. producto-capitatus sGP | 59 | Sc. pectinatus mGP | 32 |
| Sc. producto-capitatus mGP | 24 | Sc. pectinatus sGP | 43 |
| Sc. producto-capitatus sSP | 15 | Sc. pectinatus mSP | 21 |
| Sc. producto-capitatus mSP | 35 | Sc. pectinatus sSP | 63 |
| Sc. rubescens sGP | 65 | Tetradesmus wiscon. mGP | 39 |
| Sc. rubescens mGP | 23 | Tetradesmus wiscon. sGP | 72 |
| Sc. rubescens sSP | 56 | Tetradesmus wiscon. mSP | 19 |
| Sc. rubescens mSP | 22 | Tetradesmus wiscon. sSP | 16 |
| Sc. accuminatus sGP | 44 | Eustigmatos magnus mSP | 0 |
| Sc. accuminatus mGP | 26 | Eustigmatos magnus sSP | 0 |
| Sc. accuminatus sSP | 57 | Eustigmatos magnus mGP | 56 |
| Sc. accuminatus mSP | 22 | Eustigmatos magnus sGP | 61 |
| UPLC/ESI-MS | FTICR-ESI-MS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | rs | [M − H]− (m/z) | RT min | Possible Compound | Rank | rs | [M − H]− (m/z) | Possible Compound | Elemental Composition | Calcd. [M − H]− (m/z) | DBE |
| 1. | 0.75 | 815.49982 815.49827 * | 1 | C43H76O12S | 815.498472 * | 5.0 | |||||
| 4. | 0.83 | 815.7333 | 7.22 | 1 | |||||||
| 7. | 0.68 | 816.50348 | Isotope p. m/z 815.50 | ||||||||
| 8. | 0.80 | 817.6666 | 7.70 | 2 | |||||||
| 9. | 0.80 | 818.5999 | 7.70 | Isotope p. m/z 817.67 | 9. | 0.64 | 794.52123 | Isotope p. m/z 793.52 | |||
| 10. | 0.64 | 795.52035 | Isotope p. m/z 793.52 | ||||||||
| 13. | 0.63 | 817.51617 817.51596 * | 2 | C43H78O12S | 817.514122* | 4.0 | |||||
| 14. | 0.63 | 819.53126 | Isotope p. m/z 817.52 | ||||||||
| 15. | 0.75 | 794.7333 | 8.86 | Isotope p. m/z 793.73 | |||||||
| 19. | 0.56 | 793.51536 793.51441 * | 3 | C41H78O12S | 793.514122* | 2.0 | |||||
| 22. | 0.74 | 817.6333 | 7.22 | Isotope p. m/z 815.73 | |||||||
| Compound | [M − H]− (m/z) | Scan Mode [m/z] | m/z Relative Intensity (%) [Fragment Ion Schema 1] |
|---|---|---|---|
| 1 | 815 | MS2 [815] MS3 [815 → 537] | 537 ([b], 100), 559 ([a1], 38) 225 ([d], 100), 207 ([e], 7), 165 ([f], 12), 283 ([c], 26) |
| 2 | 817 | MS2 [817] MS3 [817 → 537] | 537 ([b]), 100), 561 ([a2], 32) 225 ([d]), 100), 165 ([f], 30), 207 ([e], 9), 283 ([c], 3) |
| 3 | 793 | MS2 [793] MS3 [793 → 537] MS4 [793 → 537 → 225] | 537 ([b], 100), 225 ([d], 20) 283 ([c], 42), 207 ([e], 53), 225 ([d], 100) 207 ([e], 52), 165 ([f], 60), 125 ([g], 100), 81 ([h], 27) |
| SQDG standard | 815 | MS2 [815] cMS3 [815 → 537] MS3 [815 → 559] | 537 ([b], 100), 559 ([a1], 90) 225 ([d], 100), 165 ([f], 58), 207, ([e],30) 225 ([d], 100), 283 ([c], 38) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hielscher-Michael, S.; Griehl, C.; Buchholz, M.; Demuth, H.-U.; Arnold, N.; Wessjohann, L.A. Natural Products from Microalgae with Potential against Alzheimer’s Disease: Sulfolipids Are Potent Glutaminyl Cyclase Inhibitors. Mar. Drugs 2016, 14, 203. https://doi.org/10.3390/md14110203
Hielscher-Michael S, Griehl C, Buchholz M, Demuth H-U, Arnold N, Wessjohann LA. Natural Products from Microalgae with Potential against Alzheimer’s Disease: Sulfolipids Are Potent Glutaminyl Cyclase Inhibitors. Marine Drugs. 2016; 14(11):203. https://doi.org/10.3390/md14110203
Chicago/Turabian StyleHielscher-Michael, Stephanie, Carola Griehl, Mirko Buchholz, Hans-Ulrich Demuth, Norbert Arnold, and Ludger A. Wessjohann. 2016. "Natural Products from Microalgae with Potential against Alzheimer’s Disease: Sulfolipids Are Potent Glutaminyl Cyclase Inhibitors" Marine Drugs 14, no. 11: 203. https://doi.org/10.3390/md14110203






