Diatom-Specific Oligosaccharide and Polysaccharide Structures Help to Unravel Biosynthetic Capabilities in Diatoms
Abstract
:1. Introduction
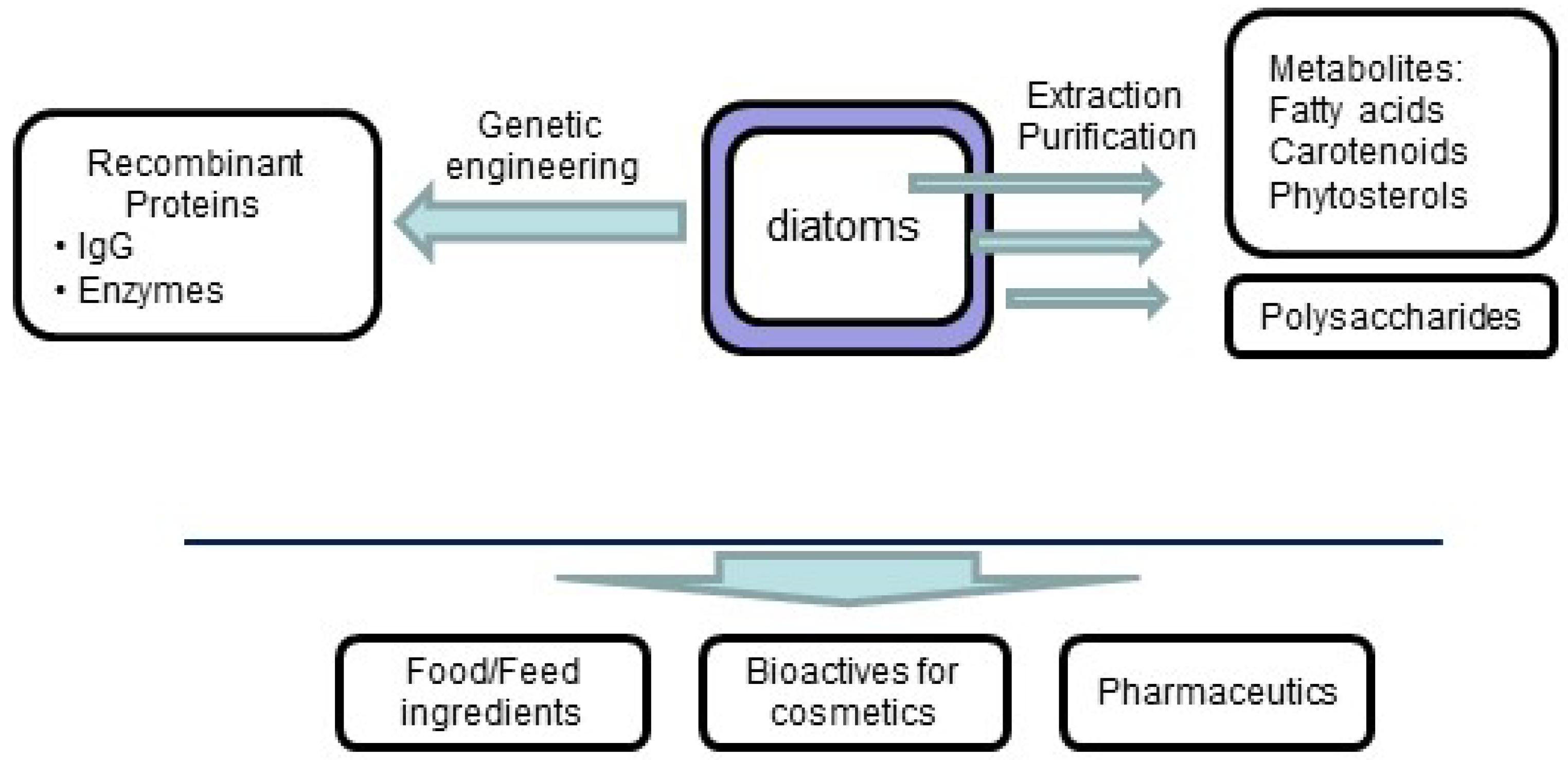
2. Monosaccharide Composition and Structures of Polysaccharides in Diatoms
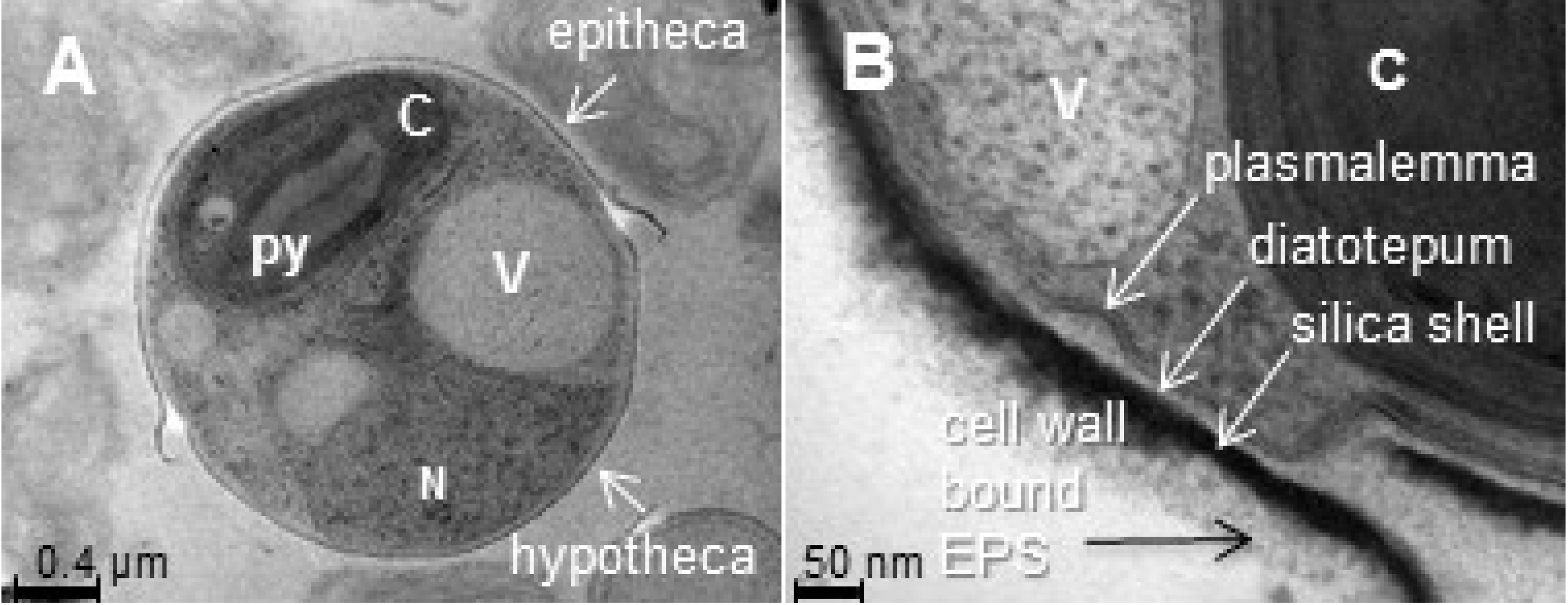
2.1. Insoluble Polysaccharides in Diatoms
2.1.1. Frustules, or Cell Wall Polysaccharides
| Monosaccharide | Ara | Fuc | Gal | Glc | Man | Rha | Rib | Xyl | 3-O-MeFuc | 2-MeGal | 3/4-MeGal | GalA | GlcA | 2-MeGlcA | ManA | 2-MeRha | 3-MeRha | 2,3-diMeRha | 3-MeXyl | 4-MeXyl | Unknown | GlcNac | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Chaeotoceros affinis | Alkali soluble fraction | 11 | 18 | 2 | 6 | 52 | — | 7 | ||||||||||||||||
| a Chaeotoceros curvisetus | 32 | 31 | 1 | 6 | 16 | 10 | 4 | |||||||||||||||||
| a Chaeotoceros decipiens | 4 | 7 | 6 | 32 | 25 | 4 | 10 | |||||||||||||||||
| a Chaeotoceros debilis | 11 | 23 | 6 | — | 22 | 23 | 15 | |||||||||||||||||
| a Chaeotoceros socialis | 18 | 16 | 3 | 9 | 23 | 11 | 8 | |||||||||||||||||
| a Thalassiosira gravida | 43 | 12 | 4 | 7 | — | 27 | 7 | |||||||||||||||||
| a Corethron hystrix | 60 | 9 | 11 | 9 | 1 | 8 | 2 | |||||||||||||||||
| b Thalassiosira weissflogii | 1.94 | 6.98 | 36 | 19.5 | 17.9 | 4 | 3.91 | 7.31 | ||||||||||||||||
| c Phaeodactylum tricornutum | 0.3 | 2.3 | 1.7 | 2.4 | 55.6 | 11.7 | — | 5.4 | nd | tr | — | 1.9 | 6.7 | 12 | nd | nd | — | nd | nd | |||||
| d Phaeodactylum tricornutum O | 7 | 3 | 8 | 18 | 28 | 9 | 1 | 8 | tr | tr | tr | |||||||||||||
| d Phaeodactylum tricornutum F | 2 | 6 | 11 | 45 | 7 | 8 | 2 | 10 | 2 | |||||||||||||||
| e Stauroneis amphioxys | Alkali insoluble fraction | — | 1 | 9 | 7 | 50 | 2 | 2 | 2 | nd | 28 | nd | nd | 2 | nd | — | — | |||||||
| d Phaeodactylum tricornutum O | 1 | 5 | 12 | 72 | 1 | tr | 1 | |||||||||||||||||
| d Phaeodactylum tricornutum F | 4 | 1 | 12 | 10 | 66 | 2 | tr | 4 | 1 | |||||||||||||||
| f Nitzschia frustulum | Insoluble organic cell walls | — | 14 | 14 | tr | 32 | 5 | — | tr | 35 | ||||||||||||||
| f Nitzschia angularis | — | 64 | 4 | 14 | 11 | tr | — | tr | 7 | |||||||||||||||
| f Asterionella socialis | — | tr | 5 | tr | 22 | tr | — | tr | 78 | |||||||||||||||
| f Cylindrotheca fusiformis | — | tr | 10 | tr | 12 | tr | — | tr | 78 | |||||||||||||||
| g Navicula pelliculosa | — | 9.2 | 9.9 | 25.9 | 48.5 | 3.8 | — | 3.1 | ||||||||||||||||
| g Melosira nummuloides | 0.3 | 25.6 | 3.7 | 0.9 | 56.8 | 1.8 | — | 10.9 | ||||||||||||||||
| g Melosira granulata | Insoluble organic cell walls | 0.6 | 0.8 | 5.4 | 46.7 | 6.5 | 1.2 | — | 38.9 | |||||||||||||||
| g Cyclotella stelligera | 2 | 0.6 | 22.4 | 43.3 | 13.6 | 0 | — | 18 | ||||||||||||||||
| g Cyclotella cryptica | — | 12.2 | 12.2 | 13 | 37.2 | 7.4 | — | 17.8 | ||||||||||||||||
| g Nitzschia brevirostris Hust. | 1.7 | 42.5 | 9.5 | 14.6 | 20.2 | 2.4 | 4.7 | 4.3 | ||||||||||||||||
| h Pinnularia viridis | tr | 1.5 | 7 | 13 | 54 | 9.5 | 11 | — | — | — | 2 | 1 | 1 | — | — | |||||||||
| h Craspedostauros australis | tr | tr | 2 | 5 | 69 | 2 | 4 | 2 | 2 | 12 | — | — | — | 2 | — | |||||||||
| h Thalassiosira pseudonana | 1 | 2 | 10 | 6 | 65 | tr | 5 | — | 10 | — | — | tr | — | — | 1 | |||||||||
| h Nitzschia navis-varingica | 2 | 1 | 3 | 7 | 64 | 2 | 1 | tr | 15 | — | — | 5 | — | — | — | |||||||||
| i Coscinodiscus radiatus | tr | 4.7 | 80.1 | 1.5 | 1.4 | tr | 12.4 | |||||||||||||||||
| i Nitzschia curvilineata | 6.5 | 45.6 | 40.3 | 1.7 | 4.4 | — | 1.4 | |||||||||||||||||
| i Amphora salina | 2.7 | 47.7 | 41.5 | 1.5 | 1.3 | tr | 5.5 | |||||||||||||||||
| i Triceratium dubium | tr | 6.4 | 67.4 | tr | 6.3 | 5.6 | 1.5 | 12.8 | ||||||||||||||||
| d Phaeodactylum tricornutum O | 3 | 3 | 11 | 61 | 12 | 2 | 4 | |||||||||||||||||
| d Phaeodactylum tricornutum F | 3 | 1 | 10 | 25 | 47 | 3 | tr | 6 | 2 |

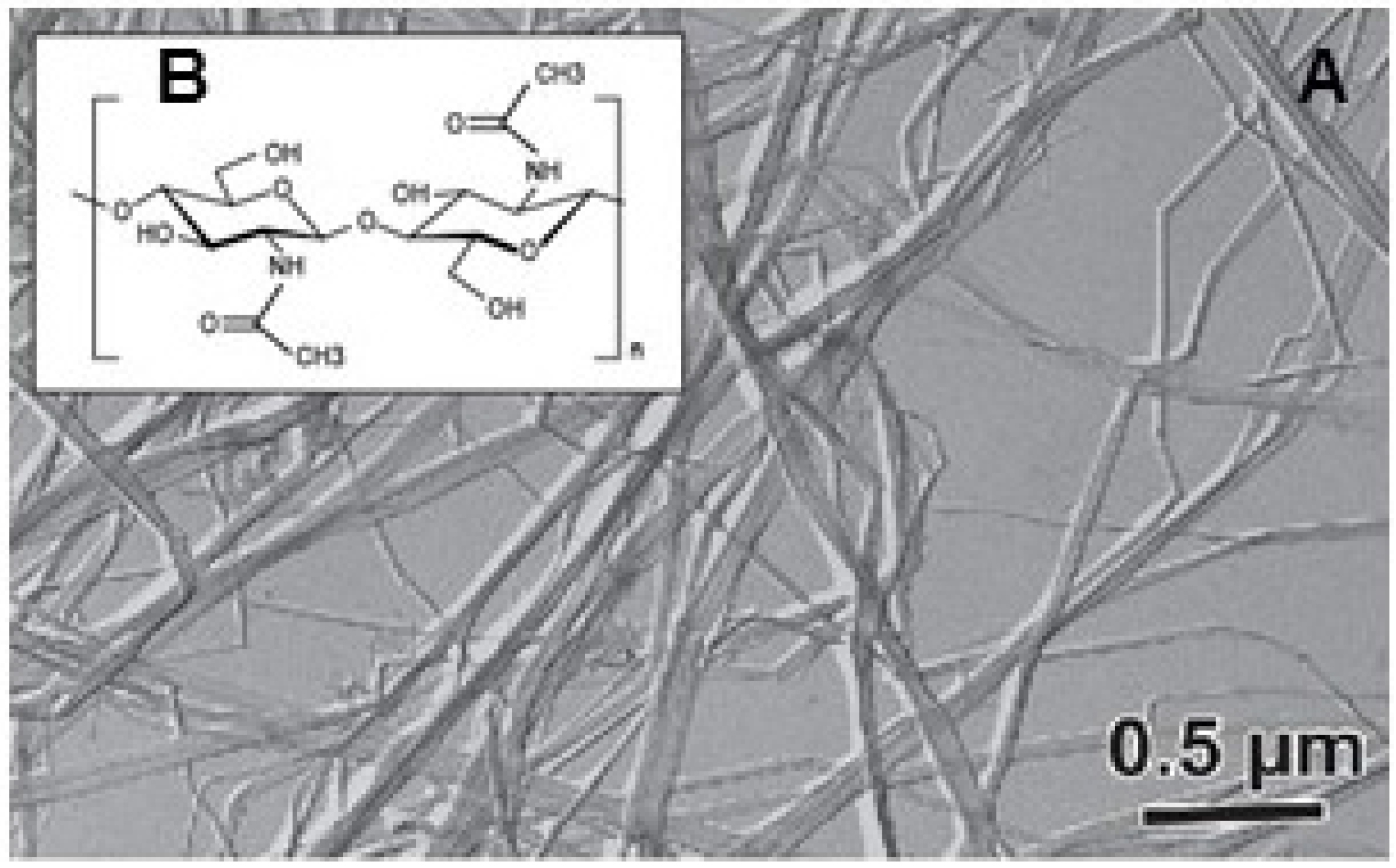
2.1.2. Chitinous Spines
2.2. Soluble Polysaccharides in Diatoms
2.2.1. Food Storage Polysaccharides

| Species | Mw/DP | Branching | Yield% (w/w) | Reference |
|---|---|---|---|---|
| Phaeodactylum tricornutum | nd | Some β(1,6) branching | 14% | [92] |
| Skeletonema costatum | 6–13 kDa | Some β(1,6) and β(1,2) branching | 32% | [96] |
| Stauroneis amphioxys | 4 kDa, DP ~24 | Some β(1,6) and β(1,2) branching | nd | [97] |
| Achnanthes longipes | nd | Small degree of β(1,6) and β(1,2) branching | nd | [98] |
| Pinnularia viridis | >10 kDa | Small degree of β(1,6) branching | nd | [99] |
| Aulacoseira baicalensis | 3–5 kDa | nd | 0.9% | [93] |
| Stephanodiscus meyerii | 40 kDa | β(1,6)/β(1,3) DB 0.053 a | 0.5% | |
| Stephanodiscus meyerii | 2–6 kDa | β(1,6)/β(1,3) DB 0.25 a | 0.4% | |
| Aulacoseira baicalensis | nd | β(1,6)/β(1,3) DB 0.11 a Mannitol detected | 0.6% | |
| Chaetoceros muelleri | DP 22–24 | β(1,6)/β(1,3) DB 0.006–0.009 | nd | [94] |
| Thalassiosira weissflogii | DP 5–13 | No branching | nd | |
| Chaetoceros debilis | 4.9 kDa, DP 30 | β(1,6) 37% of total residue | 10% | [100] |
2.2.2. Exopolysaccharides
| Species | Monosaccharides | Sulfate a (wt%) | Linkages | Reference |
|---|---|---|---|---|
| Amphora sp. F1 | GlcA(1.6)/Gal(1.1)/Fuc(1) b | 9.7 | nd c | [113] |
| Amphora sp. F2 | GlcA(2.8)/Fuc(1)/Gal(0.8) | 18.2 | nd | |
| Amphora holsatica | UA/Rha/Fuc/Glc/Xyl/Ara d | nd | nd | [114] |
| Amphora rostrata | Fuc(41)/Gal(32)/UA(23)/Man(9)/Rha(8) | 10 | nd | [115] |
| Asterionella socialis e | Rha(70)/Man(7)/2 Unk(23) | nd | nd | [69] |
| Chaetoceros affinis e | Fuc(39)/Rha(35)/Gal(26) | 8.7 | t-Fuc f, 2,3-Fuc, 3,4-Fuc, 3-Fuc/2-Rha, t-Rha, 3-Rha, 3,4-Rha/3-Gal, t-Gal, 4-Gal f | [116,117] |
| Chaetoceros curvisetus e | Fuc(35)/Gal(10)/Rha(3) | 7 | 2-Fuc f, t-Fuc f, 2,3-Fuc, 3-Fuc, 2,3-Fuc f, 3,4-Fuc, 3,5-Fuc f/3-Gal, 2,3-Gal, t-Gal/2-Rha, t-Rha | [118] |
| Chaetoceros debilis b | Fuc(30)/Gal(29)/Rha(17)/Man(10)/Xyl(9)/Glc(5) | nd | nd | [62] |
| Chaetoceros decipiens e | Rha(34)/Fuc(32)/Gal(17)/Man(7)/Xyl(5)/Glc(5) | nd | nd | |
| Coscinodiscus nobilis b | Fuc(34)/Man(19)/Glc(16)/Rha(15)/GlcA(9)/Xyl(6) | 16.7 | 3-Fuc/6-Man/3-Glc/2-Rha/t-Xyl with Fuc and Rha branched or sulfated | [119] |
| Cylindrotheca closterium | Xyl(46)/Glc(23)/Rha(15)/Gal(12)/Man(4)/UA(5) | 0 | nd | [107] |
| Cylindrotheca fusiformis | Gal(38)/Glc(26)/Xyl(13)/Rha(13)/UA(7)/Man(5) | 31 | nd | [108] |
| Cyclotella nana e | Rha(33)/Gal(14)/Glc(11)/Man(10)/Rib(8)/Xyl(7)/2 Unk(17)/GlcA(?) | nd | nd | [69] |
| Melosira nummuloides | UA/Rha/Fuc/Glc/Xyl/Ara/Gal d | nd | nd | [114] |
| Navicula directa | UA/Rha/Fuc/Glc/Ara/Gal/Xyl d | nd | nd | |
| Navicula incerta e | Rha(33)/Fuc(20)/Man(10)/Xyl(9)/Gal(8)/GlcA(?)/3 Unk(20) | nd | nd | [69] |
| Navicula salinarum | Glc(41)/Xyl(20)/Gal(19)/Man(14)/Rha(5)/UA(21) | 6.3 | nd | [107] |
| Navicula subinflata | Glc(94)/UA(9) | 9.6 | nd | [120] |
| Nitzschia angularis e | Rha(20)/Gal(17)/Fuc(16)/Ara(8)/Man(7)/Xyl(7)/GlcA(?)/2 Unk(25)/ | nd | nd | [69] |
| Nitzschia frustulum e | Man(34)/Rha(24)/Gal(8)/GlcA(?)/2 Unk(34) | 9 | nd | |
| Pinnularia viridis | Rha(29)/Gal(23.5)/Xyl(17)/Glc(7.5)/Man(6.5)/Fuc(6) | nd | 3-Rha, 3,4-Rha, 2,3-Rha, 2-Rha, t-Rha/ 3-Gal, 3,6-Gal/t-Xyl, 2,4-Xyl, 4-Xyl/4-Glc/4-Man, t-Man/t-Fuc, 2-Fuc | [99] |
| Thalassiosira sp. F1 | Man(51)/Rha(19)/Fuc(8)/Xyl(6) | nd | t-Man, 4,6-Man, 4-Man/3-Rha, 2-Rha/3-Fuc, t-Fuc/t-Xyl, 2-Xyl | [121] |
| Thalassiosira sp. F2 | Man(57)/Xyl(19)/GlcA(6)/GalA(5) | nd | 4-Man, t-Man, 2-Man/4-Xyl, t-Xyl/t-GlcA/t-GalA |
3. Structures and Biosynthesis of Protein N-Glycans in Diatoms

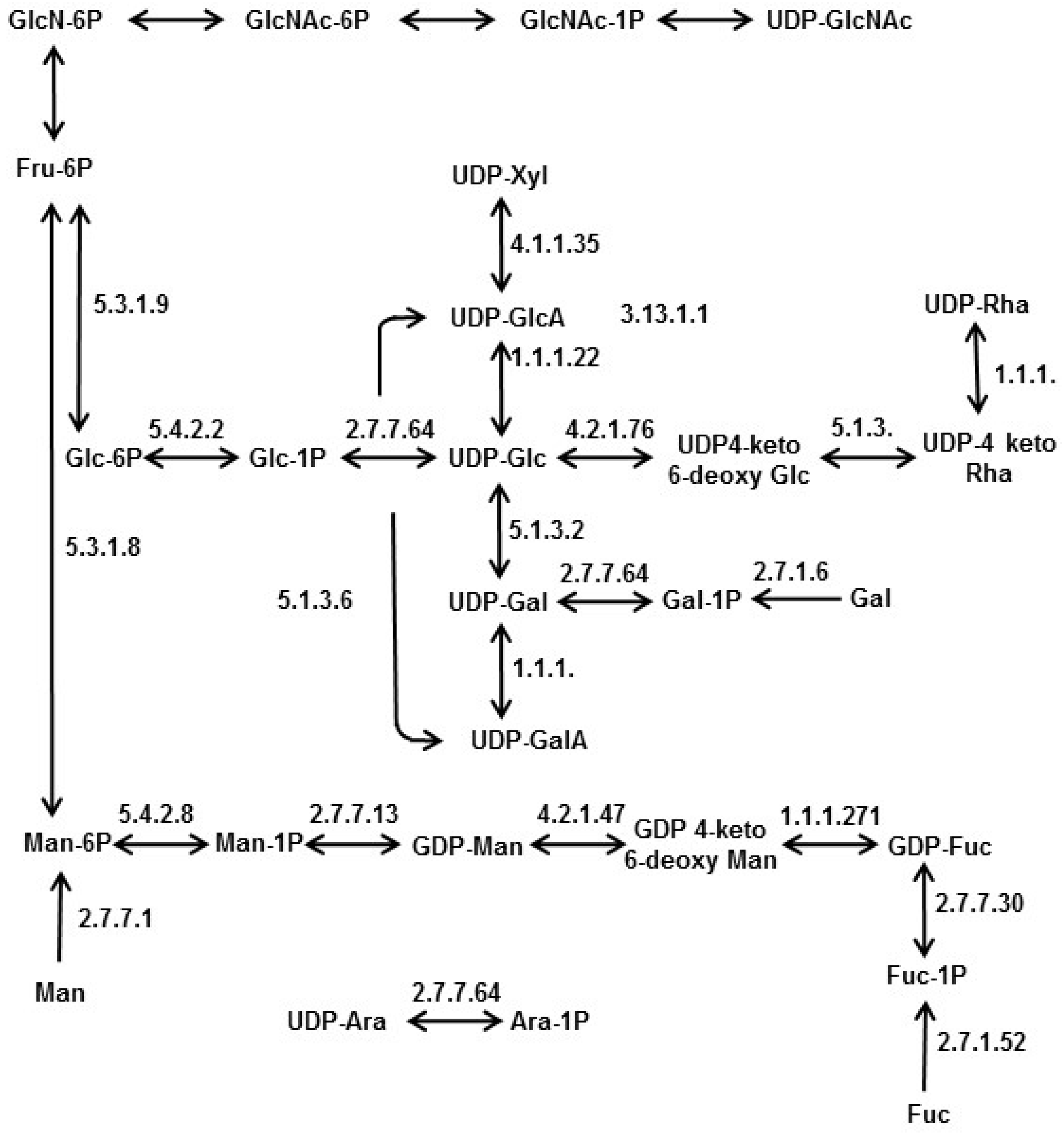
4. Nucleotide Sugar Biosynthesis in Diatoms
5. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Norton, T.A.; Melkonian, M.; Andersen, R.A. Algal biodiversity. Phycologia 1996, 35, 308–326. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Barber, R.T.; Smetacek, V. Biogeochemical controls and feedbacks on ocean primary production. Science 1998, 281, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.C. Potential role of large oceanic diatoms in new primary production. Deep Sea Res. Part Oceanogr. Res. Pap. 1993, 40, 159–168. [Google Scholar] [CrossRef]
- Caldwell, G.S. The influence of bioactive oxylipins from marine diatoms on invertebrate reproduction and development. Mar. Drugs 2009, 7, 367–400. [Google Scholar] [CrossRef] [PubMed]
- Tréguer, P.; Nelson, D.M.; van Bennekom, A.J.; de Master, D.J.; Leynaert, A.; Quéguiner, B. The silica balance in the world ocean: A reestimate. Science 1995, 268, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M.; Tréguer, P.; Brzezinski, M.A.; Leynaert, A.; Quéguiner, B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Hutchins, D.A.; Bruland, K.W. Iron-limited diatom growth and Si: N uptake ratios in a coastal upwelling regime. Nature 1998, 393, 561–564. [Google Scholar] [CrossRef]
- Kemp, A.E.S.; Pearce, R.B.; Grigorov, I.; Rance, J.; Lange, C.B.; Quilty, P.; Salter, I. Production of giant marine diatoms and their export at oceanic frontal zones: Implications for Si and C flux from stratified oceans: Giant marine diatoms and their export. Glob. Biogeochem. Cycles 2006, 20. [Google Scholar] [CrossRef]
- Allen, A.E.; LaRoche, J.; Maheswari, U.; Lommer, M.; Schauer, N.; Lopez, P.J.; Finazzi, G.; Fernie, A.R.; Bowler, C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 2008, 105, 10438–10443. [Google Scholar] [CrossRef] [PubMed]
- Sutak, R.; Botebol, H.; Blaiseau, P.-L.; Leger, T.; Bouget, F.-Y.; Camadro, J.-M.; Lesuisse, E. A Comparative Study of Iron Uptake Mechanisms in Marine Microalgae: Iron Binding at the Cell Surface Is a Critical Step. Plant Physiol. 2012, 160, 2271–2284. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Iron acquisition and allocation in stramenopile algae. J. Exp. Bot. 2013, 64, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Collos, Y. Transient situations in nitrate assimilation by marine diatoms. IV. Non-linear phenomena and the estimation of the maximum uptake rate. J. Plankton Res. 1983, 5, 677–691. [Google Scholar] [CrossRef]
- Waser, N.A.D.; Harrison, P.J.; Nielsen, B.; Calvert, S.E.; Turpin, D.H. Nitrogen isotope fractionation during the uptake and assimilation of nitrate, nitrite, ammonium, and urea by a marine diatom. Limnol. Oceanogr. 1998, 43, 215–224. [Google Scholar] [CrossRef]
- Villareal, T.A. Division cycles in the nitrogen-fixing Rhizosolenia (Bacillariophyceae)-Richelia (Nostocaceae) symbiosis. Br. Phycol. J. 1989, 24, 357–365. [Google Scholar] [CrossRef]
- Weber, T.; Deutsch, C. Oceanic nitrogen reservoir regulated by plankton diversity and ocean circulation. Nature 2012, 489, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Craggs, R.J.; McAuley, P.J.; Smith, V.J. Wastewater nutrient removal by marine microalgae grown on a corrugated raceway. Water Res. 1997, 31, 1701–1707. [Google Scholar] [CrossRef]
- Lopez, P.J.; Desclés, J.; Allen, A.E.; Bowler, C. Prospects in diatom research. Curr. Opin. Biotechnol. 2005, 16, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, T.; Robert, J.-M. Diatom cultivation and biotechnologically relevant products. Part II: Current and putative products. Appl. Microbiol. Biotechnol. 2003, 60, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Herlory, O.; Bonzom, J.-M.; Gilbin, R.; Frelon, S.; Fayolle, S.; Delmas, F.; Coste, M. Use of diatom assemblages as biomonitor of the impact of treated uranium mining effluent discharge on a stream: Case study of the Ritord watershed (Center-West France). Ecotoxicology 2013, 22, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Bræk, G.S.; Jensen, A.; Mohus, Å. Heavy metal tolerance of marine phytoplankton. III. Combined effects of copper and zinc ions on cultures of four common species. J. Exp. Mar. Biol. Ecol. 1976, 25, 37–50. [Google Scholar]
- Morelli, E.; Pratesi, E. Production of phytochelatins in the marine diatom Phaeodactylum tricornutum in response to copper and cadmium exposure. Bull. Environ. Contam. Toxicol. 1997, 59, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Pistocchi, R.; Mormile, M.A.; Guerrini, F.; Isani, G.; Boni, L. Increased production of extra- and intracellular metal-ligands in phytoplankton exposed to copper and cadmium. J. Appl. Phycol. 2000, 12, 469–477. [Google Scholar] [CrossRef]
- Parkinson, J.; Gordon, R. Beyond micromachining: The potential of diatoms. Trends Biotechnol. 1999, 17, 190–196. [Google Scholar] [CrossRef]
- Drum, R.W.; Gordon, R. Star Trek replicators and diatom nanotechnology. Trends Biotechnol. 2003, 21, 325–328. [Google Scholar] [CrossRef]
- Bozarth, A.; Maier, U.-G.; Zauner, S. Diatoms in biotechnology: Modern tools and applications. Appl. Microbiol. Biotechnol. 2009, 82, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Jamali, A.A.; Akbari, F.; Ghorakhlu, M.M.; de la Guardia, M.; Khosroushahi, A.Y. Applications of diatoms as potential microalgae in nanobiotechnology. BioImpacts 2012, 2, 83–89. [Google Scholar] [PubMed]
- Muller-Feuga, A. The role of microalgae in aquaculture: Situation and trends. J. Appl. Phycol. 2000, 12, 527–534. [Google Scholar] [CrossRef]
- Becker, E.W. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Y.; Lu, Y.; Zheng, J.-W.; Yang, W.-D.; Liu, J.-S. Biochemical and genetic engineering of diatoms for polyunsaturated fatty acid biosynthesis. Mar. Drugs 2014, 12, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Kroth, P. Molecular biology and the biotechnological potential of diatoms. In Transgenic Microalgae as Green Cell Factories; Springer: New York, NY, USA, 2007; pp. 23–33. [Google Scholar]
- Peng, J.; Yuan, J.-P.; Wu, C.-F.; Wang, J.-H. Fucoxanthin, a marine carotenoid present in brown seaweeds and diatoms: Metabolism and bioactivities relevant to human health. Mar. Drugs 2011, 9, 1806–1828. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jung, Y.-J.; Kwon, O.-N.; Cha, K.; Um, B.-H.; Chung, D.; Pan, C.-H. A potential commercial source of fucoxanthin extracted from the microalga Phaeodactylum tricornutum. Appl. Biochem. Biotechnol. 2012, 166, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Moreau, D.; Tomasoni, C.; Jacquot, C.; Kaas, R.; le Guedes, R.; Cadoret, J.-P.; Muller-Feuga, A.; Kontiza, I.; Vagias, C.; Roussis, V.; et al. Cultivated microalgae and the carotenoid fucoxanthin from Odontella aurita as potent anti-proliferative agents in bronchopulmonary and epithelial cell lines. Environ. Toxicol. Pharmacol. 2006, 22, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Dalmo, R.; Martinsen, B.; Horsberg, T.; Ramstad, A.; Syvertsen, C.; Seljelid, R.; Ingebrigtsen, K. Prophylactic effect of β(1,3)-d-glucan (laminaran) against experimental Aeromonas salmonicida and Vibrio salmonicida infections. J. Fish Dis. 1998, 21, 459–462. [Google Scholar] [CrossRef]
- Sakai, M. Current research status of fish immunostimulants. Aquaculture 1999, 172, 63–92. [Google Scholar] [CrossRef]
- Morales-Lange, B.; Bethke, J.; Schmitt, P.; Mercado, L. Phenotypical parameters as a tool to evaluate the immunostimulatory effects of laminarin in Oncorhynchus mykiss. Aquac. Res. 2014. [Google Scholar] [CrossRef]
- Skjermo, J.; Størseth, T.R.; Hansen, K.; Handå, A.; Øie, G. Evaluation of β-(1 → 3,1 → 6)-glucans and High-M alginate used as immunostimulatory dietary supplement during first feeding and weaning of Atlantic cod (Gadus morhua L.). Aquaculture 2006, 261, 1088–1101. [Google Scholar] [CrossRef]
- Kusaikin, M.; Ermakova, S.; Shevchenko, N.; Isakov, V.; Gorshkov, A.; Vereshchagin, A.; Grachev, M.; Zvyagintseva, T. Structural characteristics and antitumor activity of a new chrysolaminaran from the diatom alga Synedra acus. Chem. Nat. Compd. 2010, 46, 1–4. [Google Scholar] [CrossRef]
- Raposo, M.; de Morais, R.; Bernardo de Morais, A. Bioactivity and applications of sulphated polysaccharides from marine microalgae. Mar. Drugs 2013, 11, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Abida, H.; Ruchaud, S.; Rios, L.; Humeau, A.; Probert, I.; de Vargas, C.; Bach, S.; Bowler, C. Bioprospecting marine plankton. Mar. Drugs 2013, 11, 4594–4611. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maier, U.G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Factories 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Lingg, N.; Zhang, P.; Song, Z.; Bardor, M. The sweet tooth of biopharmaceuticals: Importance of recombinant protein glycosylation analysis. Biotechnol. J. 2012, 7, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Van Beers, M.M.C.; Bardor, M. Minimizing immunogenicity of biopharmaceuticals by controlling critical quality attributes of proteins. Biotechnol. J. 2012, 7, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Gotté, M.; Plancot, B.; Lerouge, P.; Bardor, M.; Driouich, A. Cell wall O-glycoproteins and N-glycoproteins: Aspects of biosynthesis and function. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, M.; O’Neill, M.A. Plant Nucleotide Sugar Formation, Interconversion, and Salvage by Sugar Recycling. Annu. Rev. Plant Biol. 2011, 62, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, K.D.; Rosowski, J.R.; Gretz, M.R.; Roemer, S.C. Diatom extracellular polymeric substances: Function, fine structure, chemistry, and physiology. J. Phycol. 1993, 29, 537–566. [Google Scholar] [CrossRef]
- Underwood, G.J.C.; Paterson, D.M. The importance of extracellular carbohydrate production by marine epipelic diatoms. Adv. Bot. Res. 2003, 40, 83–240. [Google Scholar]
- Chiovitti, A.; Higgins, M.J.; Harper, R.E.; Wetherbee, R.; Bacic, A. The complex polysaccharides of the raphid diatom Pinnularia viridis (Bacillariophyceae). J. Phycol. 2003, 39, 543–554. [Google Scholar] [CrossRef]
- Chiovitti, A.; Molino, P.; Crawford, S.A.; Teng, R.; Spurck, T.; Wetherbee, R. The glucans extracted with warm water from diatoms are mainly derived from intracellular chrysolaminaran and not extracellular polysaccharides. Eur. J. Phycol. 2004, 39, 117–128. [Google Scholar] [CrossRef]
- Chiovitti, A.; Harper, R.E.; Willis, A.; Bacic, A.; Mulvaney, P.; Wetherbee, R. Variations in the substituted 3-linked mannans closely associated with the silicified walls of diatoms: Substituted mannans of diatoms. J. Phycol. 2005, 41, 1154–1161. [Google Scholar] [CrossRef]
- Tesson, B.; Hildebrand, M. Characterization and localization of insoluble organic matrices associated with diatom cell walls: Insight into their roles during cell wall formation. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Volcani, B. 3,4-Dihydroxyproline: A new amino acid in diatom cell walls. Science 1969, 164, 1400–1401. [Google Scholar] [CrossRef] [PubMed]
- Hecky, R.E.; Mopper, K.; Kilham, P.; Degens, E.T. The amino acid and sugar composition of diatom cell-walls. Mar. Biol. 1973, 19, 323–331. [Google Scholar] [CrossRef]
- Swift, D.M.; Wheeler, A. Evidence of an organic matrix from diatom biosilica. J. Phycol. 1992, 28, 202–209. [Google Scholar] [CrossRef]
- Kröger, N. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 1999, 286, 1129–1132. [Google Scholar] [PubMed]
- Volcani, B.E. Cell wall formation in diatoms: Morphogenesis and biochemistry. In Silicon and Siliceous Structures in Biological Systems; Simpson, T., Volcani, B., Eds.; Springer: New York, NY, USA, 1981; pp. 157–200. [Google Scholar]
- Kröger, N.; Poulsen, N. Diatoms from cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 2008, 42, 83–107. [Google Scholar] [CrossRef] [PubMed]
- Kates, M.; Volcani, B. Studies on the biochemistry and fine structure of silica shell formation in diatoms. Lipid components of the cell walls. Z. Pflanzenphysiol. 1968, 60, 19–29. [Google Scholar]
- Tesson, B.; Genet, M.J.; Fernandez, V.; Degand, S.; Rouxhet, P.G.; Martin-Jézéquel, V. Surface chemical composition of diatoms. ChemBioChem 2009, 10, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Haug, A.; Myklestad, S. Polysaccharides of marine diatoms with special reference to Chaetoceros species. Mar. Biol. 1976, 34, 217–222. [Google Scholar] [CrossRef]
- Cowie, G.L.; Hedges, J.I. Digestion and alteration of the biochemical constituents of a diatom (Thalassiosira weisflogii) ingested by an herbivorous zooplankton (Calanus pacificus). Limnol. Oceanogr. 1996, 41, 581–594. [Google Scholar] [CrossRef]
- Coombs, J.; Volcani, B. Studies on the biochemistry and fine structure of silica shell formation in diatoms. Planta 1968, 80, 264–279. [Google Scholar] [CrossRef]
- Ford, C.W.; Percival, E. Carbohydrates of Phaeodactylum tricornutum. Part II. A sulphated glucuronomannan. J. Chem. Soc. 1965, 7042–7046. [Google Scholar] [CrossRef]
- Abdullahi, A.S.; Underwood, G.J.C.; Gretz, M.R. Extracellular matrix assembly in diatoms (Bacillariophyceae). V. Environmental effects on polysaccharide synthesis in the model diatom, Phaeodactylum tricornutum. J. Phycol. 2006, 42, 363–378. [Google Scholar] [CrossRef]
- Willis, A.; Chiovitti, A.; Dugdale, T.M.; Wetherbee, R. Characterization of the extracellular matrix of Phaeodactylum tricornutum (Bacillariophyceae): Structure, composition, and adhesive characteristics. J. Phycol. 2013, 49, 937–949. [Google Scholar]
- McConville, M.J.; Wetherbee, R.; Bacic, A. Subcellular location and composition of the wall and secreted extracellular sulphated polysaccharides/proteoglycans of the diatom Stauroneis amphioxys Gregory. Protoplasma 1999, 206, 188–200. [Google Scholar] [CrossRef]
- Allan, G.G.; Lewin, J.; Johnson, P.G. Marine polymers. IV Diatom polysaccharides. Bot. Mar. 1972, 15, 102–108. [Google Scholar] [CrossRef]
- Dweltz, N.; Colvin, J.R.; McInnes, A. Studies on chitan (β-(1 → 4)-linked 2-acetamido-2-deoxy-d-glucan) fibers of the diatom Thalassiosira fluviatilis, Hustedt. III. The structure of chitan from X-ray diffraction and electron microscope observations. Can. J. Chem. 1968, 46, 1513–1521. [Google Scholar] [CrossRef]
- Blackwell, J.; Parker, K.D.; Rudall, K.M. Letter to the Editor: Chitin fibres of the diatoms Thalassiosira fluviatilis and Cyclotella cryptica. J. Mol. Biol. 1967, 28, 383–385. [Google Scholar] [CrossRef]
- Lindsay, G.J.; Gooday, G.W. Action of chitinase on spines of the diatom Thalassiosira fluviatilis. Carbohydr. Polym. 1985, 5, 131–140. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Noishiki, Y.; Wada, M. X-ray Structure of Anhydrous β-Chitin at 1 Å Resolution. Macromolecules 2011, 44, 950–957. [Google Scholar] [CrossRef]
- Sawada, D.; Nishiyama, Y.; Langan, P.; Forsyth, V.T.; Kimura, S.; Wada, M. Water in crystalline fibers of dihydrate β-chitin results in unexpected absence of intramolecular hydrogen bonding. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.; Parker, K.D.; Rudall, K.M. Chitin in pogonophore tubes. J. Mar. Biol. Assoc. UK 1965, 45, 659–661. [Google Scholar] [CrossRef]
- Gaill, F.; Persson, J.; Sugiyama, J.; Vuong, R.; Chanzy, H. The chitin system in the tubes of deep sea hydrothermal vent worms. J. Struct. Biol. 1992, 109, 116–128. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kimura, S.; Wada, M. Electron diffraction and high-resolution imaging on highly-crystalline β-chitin microfibril. J. Struct. Biol. 2011, 176, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Herth, W. A special chitin-fibril-synthesizing apparatus in the centric diatom Cyclotella. Naturwissenschaften 1978, 65, 260–261. [Google Scholar] [CrossRef]
- Herth, W. The site of β-chitin fibril formation in centric diatoms. II. The chitin-forming cytoplasmic structures. J. Ultrastruct. Res. 1979, 68, 16–27. [Google Scholar] [CrossRef]
- Herth, W.; Barthlott, W. The site of β-chitin fibril formation in centric diatoms. I. Pores and fibril formation. J. Ultrastruct. Res. 1979, 68, 6–15. [Google Scholar] [CrossRef]
- Shillito, B.; Koster, A.J.; Walz, J.; Baumeister, W. Electron tomographic reconstruction of plastic-embedded organelles involved in the chitin secretion process. Biol. Cell 1996, 88, 5–13. [Google Scholar] [CrossRef]
- Ravaux, J.; Shillito, B.; Gaill, F.; Gay, L.; Voss-Foucart, M.-F.; Childress, J. Tube synthesis and growth processes in the hydrothermal vent tube-worm Riftia pachyptila. Cah. Biol. Mar. 1998, 39, 325–326. [Google Scholar]
- Sugiyama, J.; Boisset, C.; Hashimoto, M.; Watanabe, T. Molecular directionality of β-chitin biosynthesis. J. Mol. Biol. 1999, 286, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Watanabe, T.; Yui, T.; Sugiyama, J. The directionality of chitin biosynthesis: A revisit. Biochem. J. 2003, 374, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Durkin, C.A.; Mock, T.; Armbrust, E.V. Chitin in diatoms and its association with the cell wall. Eukaryot. Cell 2009, 8, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Tesson, B.; Masse, S.; Laurent, G.; Maquet, J.; Livage, J.; Martin-Jézéquel, V.; Coradin, T. Contribution of multi-nuclear solid state NMR to the characterization of the Thalassiosira pseudonana diatom cell wall. Anal. Bioanal. Chem. 2008, 390, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Morin, L.G.; Smucker, R.A.; Herth, W. Effects of two chitin synthesis inhibitors on Thalassiosira fluviatilis and Cyclotella cryptica. FEMS Microbiol. Lett. 1986, 37, 263–268. [Google Scholar] [CrossRef]
- Quillet, M.; Combes, R. Sur la nature chimique de la leucosine, polysaccharide de réserve caractéristique des Chrysophycées, extraite d’Hydrudus foetidus. C. R. Hebd. Séances Acad. Sci. 1955, 240, 1001–1003. [Google Scholar]
- Waterkeyn, L.; Bienfait, A. Localization and function of β(1,3)-glucans (callose and chrysolaminarin) in Pinnularia genus (Diatoms). Cellule (Belg.) 1987, 74, 199–226. [Google Scholar]
- Myklestad, S. Production of carbohydrates by marine planktonic diatoms. I. Comparison of nine different species in culture. J. Exp. Mar. Biol. Ecol. 1974, 15, 261–274. [Google Scholar] [CrossRef]
- Beattie, A.; Hirst, E.L.; Percival, E. Studies on the metabolism of the Chrysophyceae. Biochem. J. 1961, 79, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.W.; Percival, E. The carbohydrates of Phaeodactylum tricornutum. Part I. Preliminary examination of the organism, and characterisation of low molecular weight material and of a glucan. J. Chem. Soc. 1965, 7035–7041. [Google Scholar] [CrossRef]
- Alekseeva, S.A.; Shevchenko, N.M.; Kusaykin, M.I.; Ponomorenko, L.P.; Isakov, V.V.; Zvyagintseva, T.N.; Likhoshvai, E.V. Polysaccharides of diatoms occurring in Lake Baikal. Appl. Biochem. Microbiol. 2005, 41, 185–191. [Google Scholar] [CrossRef]
- Størseth, T.R.; Hansen, K.; Reitan, K.I.; Skjermo, J. Structural characterization of β-d-(1 → 3)-glucans from different growth phases of the marine diatoms Chaetoceros mülleri and Thalassiosira weissflogii. Carbohydr. Res. 2005, 340, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-T.; Kim, E.-H.; Cheong, C.; Williams, D.L.; Kim, C.-W.; Lim, S.-T. Structural characterization of β-d-(1 → 3, 1 → 6)-linked glucans using NMR spectroscopy. Carbohydr. Res. 2000, 328, 331–341. [Google Scholar] [PubMed]
- Paulsen, B.S.; Myklestad, S. Structural studies of the reserve glucan produced by the marine diatom Skeletonema costatum (Grev.) Cleve. Carbohydr. Res. 1978, 62, 386–388. [Google Scholar] [CrossRef]
- McConville, M.J.; Bacic, A.; Clarke, A.E. Structural studies of chrysolaminaran from the ice diatom Stauroneis amphioxys (Gregory). Carbohydr. Res. 1986, 153, 330–333. [Google Scholar] [CrossRef]
- Wustman, B.A.; Gretz, M.R.; Hoagland, K.D. Extracellular matrix assembly in diatoms (Bacillariophyceae). I. A model of adhesives based on chemical characterization and localization of polysaccharides from the marine diatom Achnanthes longipes and other diatoms. Plant Physiol. 1997, 113, 1059–1069. [Google Scholar] [PubMed]
- Chiovitti, A.; Bacic, A.; Burke, J.; Wetherbee, R. Heterogeneous xylose-rich glycans are associated with extracellular glycoproteins from the biofouling diatom Craspedostauros australis (Bacillariophyceae). Eur. J. Phycol. 2003, 38, 351–360. [Google Scholar] [CrossRef]
- Størseth, T.R.; Kirkvold, S.; Skjermo, J.; Reitan, K.I. A branched β-d-(1 → 3, 1 → 6)-glucan from the marine diatom Chaetoceros debilis (Bacillariophyceae) characterized by NMR. Carbohydr. Res. 2006, 341, 2108–2114. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, Y.-J.; Kim, H.J.; Kim, Y.-S.; Park, W. Immunostimulatory effect of laminarin on RAW 264.7 mouse macrophages. Molecules 2012, 17, 5404–5411. [Google Scholar] [CrossRef] [PubMed]
- De Brouwer, J.; Wolfstein, K.; Stal, L.J. Physical characterization and diel dynamics of different fractions of extracellular polysaccharides in an axenic culture of a benthic diatom. Eur. J. Phycol. 2002, 37, 37–44. [Google Scholar] [CrossRef]
- Underwood, G.J.C.; Boulcott, M.; Raines, C.A.; Waldron, K. Environmental effects on exopolymer production by marine benthic diatoms: Dynamics, changes in composition, and pathways of production: Exopolymer production by diatoms. J. Phycol. 2004, 40, 293–304. [Google Scholar] [CrossRef]
- Bellinger, B.; Abdullahi, A.; Gretz, M.; Underwood, G. Biofilm polymers: Relationship between carbohydrate biopolymers from estuarine mudflats and unialgal cultures of benthic diatoms. Aquat. Microb. Ecol. 2005, 38, 169–180. [Google Scholar] [CrossRef]
- Hanlon, A.R.M.; Bellinger, B.; Haynes, K.; Xiao, G.; Hofmann, T.A.; Gretz, M.R.; Ball, A.S.; Osborn, A.M.; Underwood, G.J.C. Dynamics of extracellular polymeric substance (EPS) production and loss in an estuarine, diatom-dominated, microalgal biofilm over a tidal emersio-immersion period. Limnol. Oceanogr. 2006, 51, 79–93. [Google Scholar] [CrossRef]
- Penna, A.; Berluti, S.; Penna, N.; Magnani, M. Influence of nutrient ratios on the in vitro extracellular polysaccharide production by marine diatoms from the Adriatic Sea. J. Plankton Res. 1999, 21, 1681–1690. [Google Scholar] [CrossRef]
- Staats, N.; de Winder, B.; Stal, L.; Mur, L. Isolation and characterization of extracellular polysaccharides from the epipelic diatoms Cylindrotheca closterium and Navicula salinarum. Eur. J. Phycol. 1999, 34, 161–169. [Google Scholar] [CrossRef]
- Magaletti, E.; Urbani, R.; Sist, P.; Ferrari, C.R.; Cicero, A.M. Abundance and chemical characterization of extracellular carbohydrates released by the marine diatom Cylindrotheca fusiformis under N-and P-limitation. Eur. J. Phycol. 2004, 39, 133–142. [Google Scholar] [CrossRef]
- Urbani, R.; Magaletti, E.; Sist, P.; Cicero, A.M. Extracellular carbohydrates released by the marine diatoms Cylindrotheca closterium, Thalassiosira pseudonana and Skeletonema costatum: Effect of P-depletion and growth status. Sci. Total Environ. 2005, 353, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.-X.; Liang, J.-R.; Gao, Y.-H.; Lo, S.C.-L.; Lee, F.W.-F.; Chen, C.-P.; Luo, C.-S.; Du, C. MALDI-TOF MS analysis of the extracellular polysaccharides released by the diatom Thalassiosira pseudonana under various nutrient conditions. J. Appl. Phycol. 2015, 27, 673–684. [Google Scholar] [CrossRef]
- Wustman, B.A.; Lind, J.; Wetherbee, R.; Gretz, M.R. Extracellular matrix assembly in diatoms (Bacillariophyceae) III. Organization of fucoglucuronogalactans within the adhesive stalks of Achnanthes longipes. Plant Physiol. 1998, 116, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, E.; Ledauphin, J.; Goux, D.; Orvain, F. Optimising extraction of extracellular polymeric substances (EPS) from benthic diatoms: Comparison of the efficiency of six EPS extraction methods. Mar. Freshw. Res. 2009, 60, 1201–1210. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, C.; Santschi, P.H. Chemical composition and 234Th (IV) binding of extracellular polymeric substances (EPS) produced by the marine diatom Amphora sp. Mar. Chem. 2008, 112, 81–92. [Google Scholar] [CrossRef]
- Leandro, S.M.; Gil, M.C.; Delgadillo, I. Partial characterisation of exopolysaccharides exudated by planktonic diatoms maintained in batch cultures. Acta Oecol. 2003, 24, S49–S55. [Google Scholar] [CrossRef]
- Khandeparker, R.D.; Bhosle, N.B. Extracellular polymeric substances of the marine fouling diatom Amphora rostrata Wm.Sm. Biofouling 2001, 17, 117–127. [Google Scholar] [CrossRef]
- Myklestad, S.; Haug, A.; Larsen, B. Production of carbohydrates by the marine diatom Chaetoceros affinis var. willei (Gran) Hustedt. II. Preliminary investigation of the extracellular polysaccharide. J. Exp. Mar. Biol. Ecol. 1972, 9, 137–144. [Google Scholar] [CrossRef]
- Smestad, B.; Haug, A.; Myklestad, S. Production of carbohydrate by the marine diatom Chaetoceros affinis var. willei (Gran) Hustedt. III. Structural studies of the extracellular polysaccharide. Acta Chem. Scand. B 1974, 28, 662–666. [Google Scholar] [CrossRef]
- Smestad, B.; Haug, A.; Myklestad, S. Structural studies of the extracellular polysaccharide produced by the diatom Chaeotoceros curvisetus Cleve. Acta Chem. Scand. B 1975, 29B, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Percival, E.; Rahman, M.A.; Weigel, H. Chemistry of the polysaccharides of the diatom Coscinodiscus nobilis. Phytochemistry 1980, 19, 809–811. [Google Scholar] [CrossRef]
- Bhosle, N.; Sawant, S.; Garg, A.; Wagh, A. Isolation and partial chemical analysis of exopolysaccharides from the marine fouling diatom Navicula subinflata. Bot. Mar. 1995, 38, 103–110. [Google Scholar] [CrossRef]
- Giroldo, D.; Vieira, A.A.H.; Paulsen, B.S. Relative increase of deoxy sugars during microbial degradation of an extracellular polysaccharide released by a tropical freshwater Thalassiosira sp. (Bacillariophyceae). J. Phycol. 2003, 39, 1109–1115. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Marth, J.D.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Symbol nomenclature for glycan representation. Proteomics 2009, 9, 5398–5399. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Aebi, M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta (BBA) Gen. Subj. 1999, 1426, 239–257. [Google Scholar] [CrossRef]
- Gil, G.-C.; Velander, W.H.; van Cott, K.E. N-glycosylation microheterogeneity and site occupancy of an Asn-X-Cys sequon in plasma-derived and recombinant protein C. Proteomics 2009, 9, 2555–2567. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, D.F.; Gnad, F.; Wiśniewski, J.R.; Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Takita, E.; Sato, T.; Kinjo, S.; Aizawa, M.; Sugiura, Y.; Hamabata, T.; Sawada, K.; Kato, K. N-glycosylation at noncanonical Asn-X-Cys sequences in plant cells. Glycobiology 2011, 21, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Weerapana, E.; Imperiali, B. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology 2006, 16, 91R–101R. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef] [PubMed]
- JGI Genome Portal, Fragilariopsis cylindrus Home. Available online: http://genome.jgi-psf.org/Fracy1/Fracy1.home.html (accessed on 24 November 2014).
- Gobler, C.J.; Berry, D.L.; Dyhrman, S.T.; Wilhelm, S.W.; Salamov, A.; Lobanov, A.V.; Zhang, Y.; Collier, J.L.; Wurch, L.L.; Kustka, A.B.; et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc. Natl. Acad. Sci. USA 2011, 108, 4352–4357. [Google Scholar] [CrossRef] [PubMed]
- Baiet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.-C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-Glycans of Phaeodactylum tricornutum diatom and functional characterization of Its N-acetylglucosaminyl transferase I enzyme. J. Biol. Chem. 2011, 286, 6152–6164. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Rivet, E.; Kiefer-Meyer, M.-C.; Vanier, G.; Ovide, C.; Burel, C.; Lerouge, P.; Bardor, M. Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Levy-Ontman, O.; Fisher, M.; Shotland, Y.; Weinstein, Y.; Tekoah, Y.; Arad, S. Genes involved in the endoplasmic reticulum N-glycosylation pathway of the red microalga Porphyridium sp.: A bioinformatic study. Int. J. Mol. Sci. 2014, 15, 2305–2326. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Evolutionary forces shaping the Golgi glycosylation machinery: Why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 1993, 3, 97–130. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, P.; Varki, A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 1999, 9, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Chrispeels, M.J. Transient N-acetylglucosamine in the biosynthesis of phytohemagglutinin: Attachment in the Golgi apparatus and removal in protein bodies. J. Cell Biol. 1984, 99, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Altmann, F.; Schwihla, H.; Staudacher, E.; Glössl, J.; März, L. Insect cells contain an unusual, membrane-bound-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J. Biol. Chem. 1995, 270, 17344–17349. [Google Scholar] [CrossRef] [PubMed]
- Kröger, N.; Bergsdorf, C.; Sumper, M. A new calcium binding glycoprotein family constitutes a major diatom cell wall component. EMBO J. 1994, 13, 4676–4683. [Google Scholar] [PubMed]
- Lind, J.L.; Heimann, K.; Miller, E.A.; van Vliet, C.; Hoogenraad, N.J.; Wetherbee, R. Substratum adhesion and gliding in a diatom are mediated by extracellular proteoglycans. Planta 1997, 203, 213–221. [Google Scholar] [CrossRef] [PubMed]
- KEGG PATHWAY Database. Available online: http://www.genome.jp/kegg/pathway.html (accessed on 24 November 2014).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gügi, B.; Le Costaouec, T.; Burel, C.; Lerouge, P.; Helbert, W.; Bardor, M. Diatom-Specific Oligosaccharide and Polysaccharide Structures Help to Unravel Biosynthetic Capabilities in Diatoms. Mar. Drugs 2015, 13, 5993-6018. https://doi.org/10.3390/md13095993
Gügi B, Le Costaouec T, Burel C, Lerouge P, Helbert W, Bardor M. Diatom-Specific Oligosaccharide and Polysaccharide Structures Help to Unravel Biosynthetic Capabilities in Diatoms. Marine Drugs. 2015; 13(9):5993-6018. https://doi.org/10.3390/md13095993
Chicago/Turabian StyleGügi, Bruno, Tinaïg Le Costaouec, Carole Burel, Patrice Lerouge, William Helbert, and Muriel Bardor. 2015. "Diatom-Specific Oligosaccharide and Polysaccharide Structures Help to Unravel Biosynthetic Capabilities in Diatoms" Marine Drugs 13, no. 9: 5993-6018. https://doi.org/10.3390/md13095993