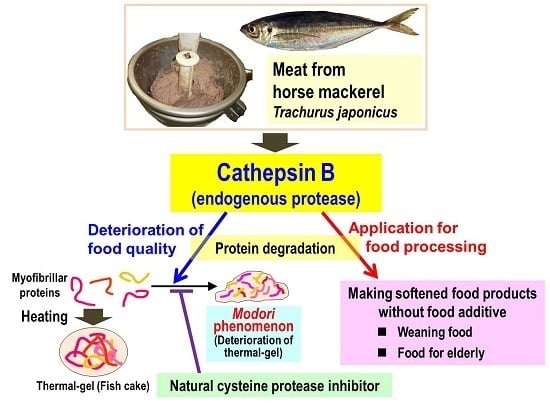
Purification and Characterization of Cathepsin B from the Muscle of Horse Mackerel Trachurus japonicus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification of Cathepsin B from Horse Mackerel Meat
| Purification Step | Total Protein (mg) | Total Activity (×10−4 U) * | Specific Activity (×10−4 U/mg) | Purity (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude enzyme | 61,827.35 | 88,272.26 | 1.43 | 1 | 100.0 |
| 50%–70% (NH4)2SO4 | 6732.00 | 26,081.32 | 3.87 | 3 | 29.5 |
| SP-Sepharose | 39.00 | 62,231.00 | 1595.67 | 1118 | 70.5 |
| Superdex 75 | 1.37 | 2512.24 | 1832.51 | 1284 | 2.8 |
| Mono S | 0.34 | 1520.14 | 4470.99 | 3132 | 1.7 |
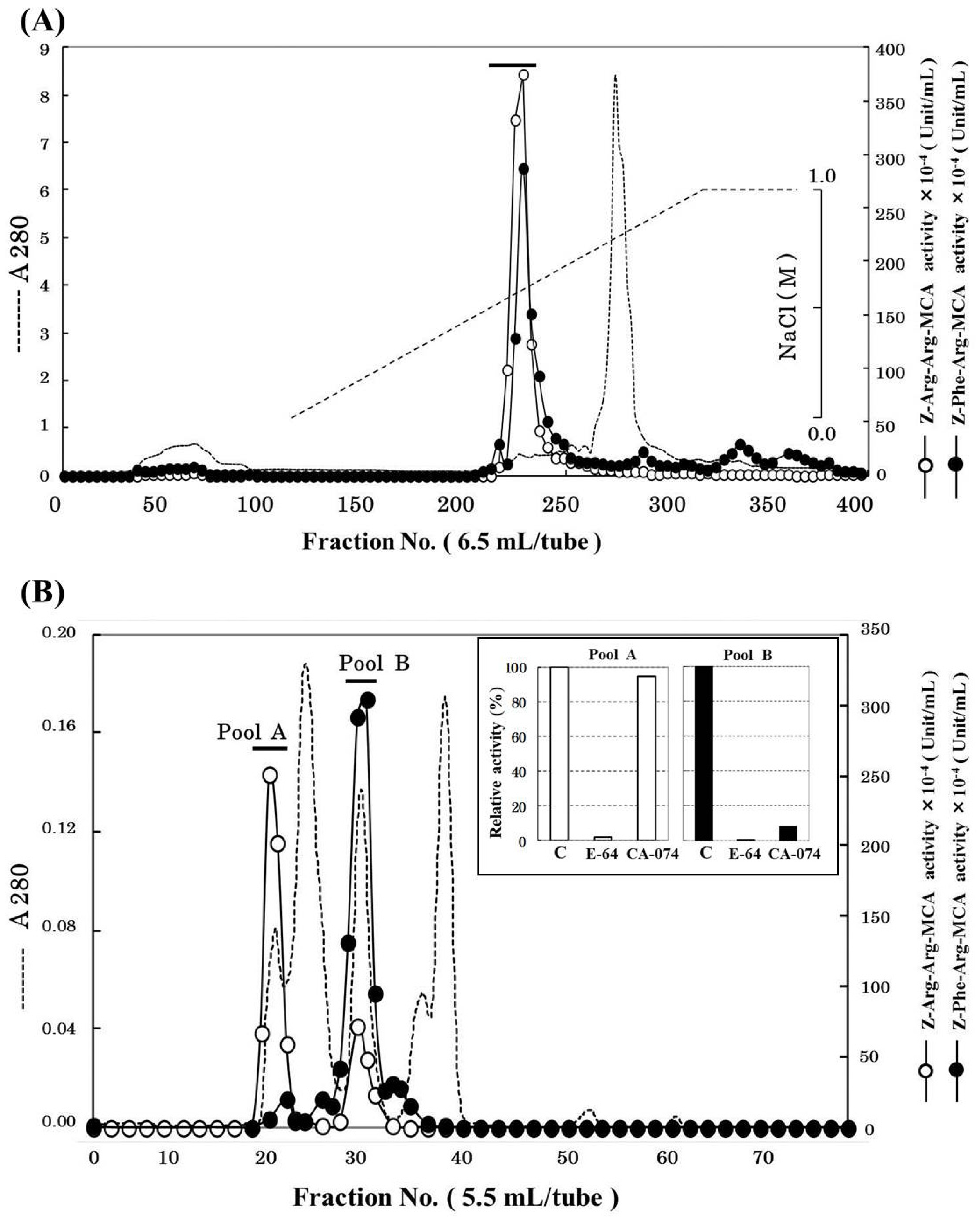

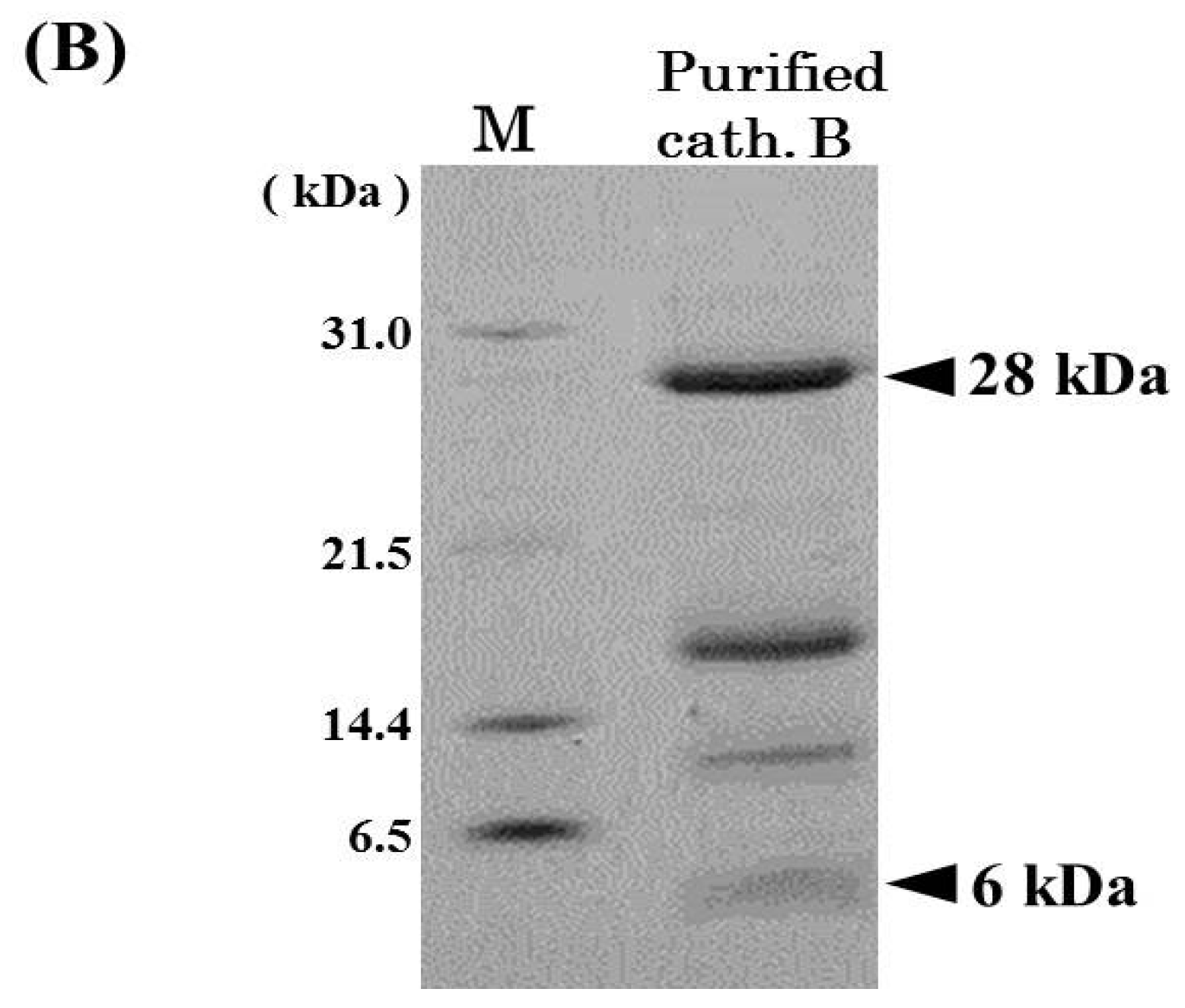
2.2. Effects of pH and Temperature on Cathepsin B Activity
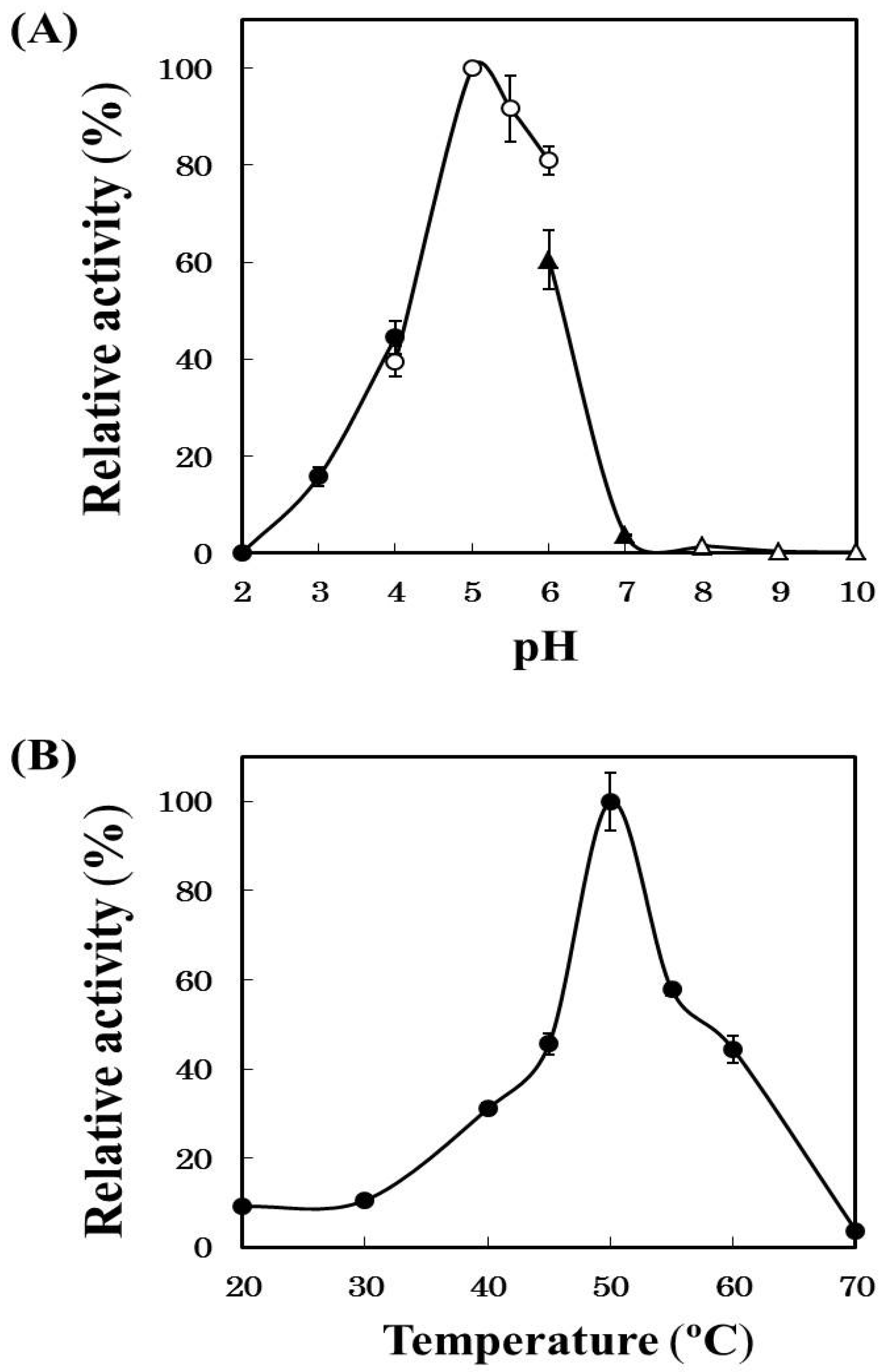
2.3. Effects of Protease Inhibitors on Cathepsin B Activity
| Reagents | Concentration (mM) | Relative Activity (%) * |
|---|---|---|
| Control | 100 | |
| E-64 | 0.01 | 1 |
| CA-074 | 0.01 | 3 |
| Chymostatin | 0.10 | 4 |
| Pefabloc SC | 1.00 | 89 |
| Pepstatin A | 0.01 | 103 |
| EDTA | 1.00 | 122 |
| 2-Mercaptoethanol | 2.00 | 175 |
2.4. Substrate Specificity of Purified Cathepsin B
| Substrates | Relative Activity (%) * |
|---|---|
| Z-Phe-Arg-MCA | 100 |
| Z-Arg-Arg-MCA | 17 |
| Arg-MCA | 0 |
| Boc-Phe-Ser-Arg-MCA | 29 |
| Boc-Val-Pro-Arg-MCA | 8 |
| Suc-Leu-Leu-Val-Tyr-MCA | 6 |
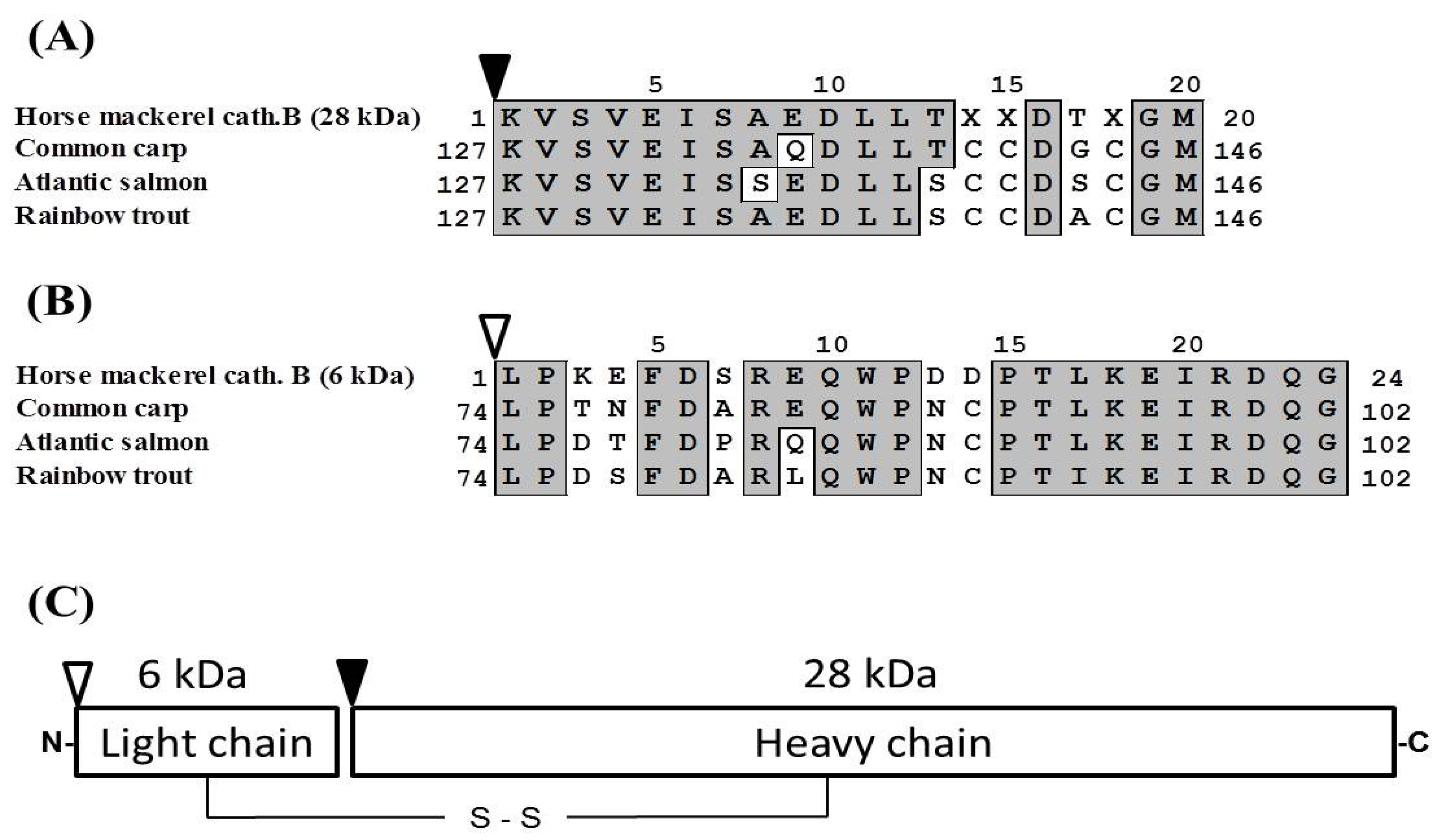
2.5. Molecular Cloning of Cathepsin B from Horse Mackerel
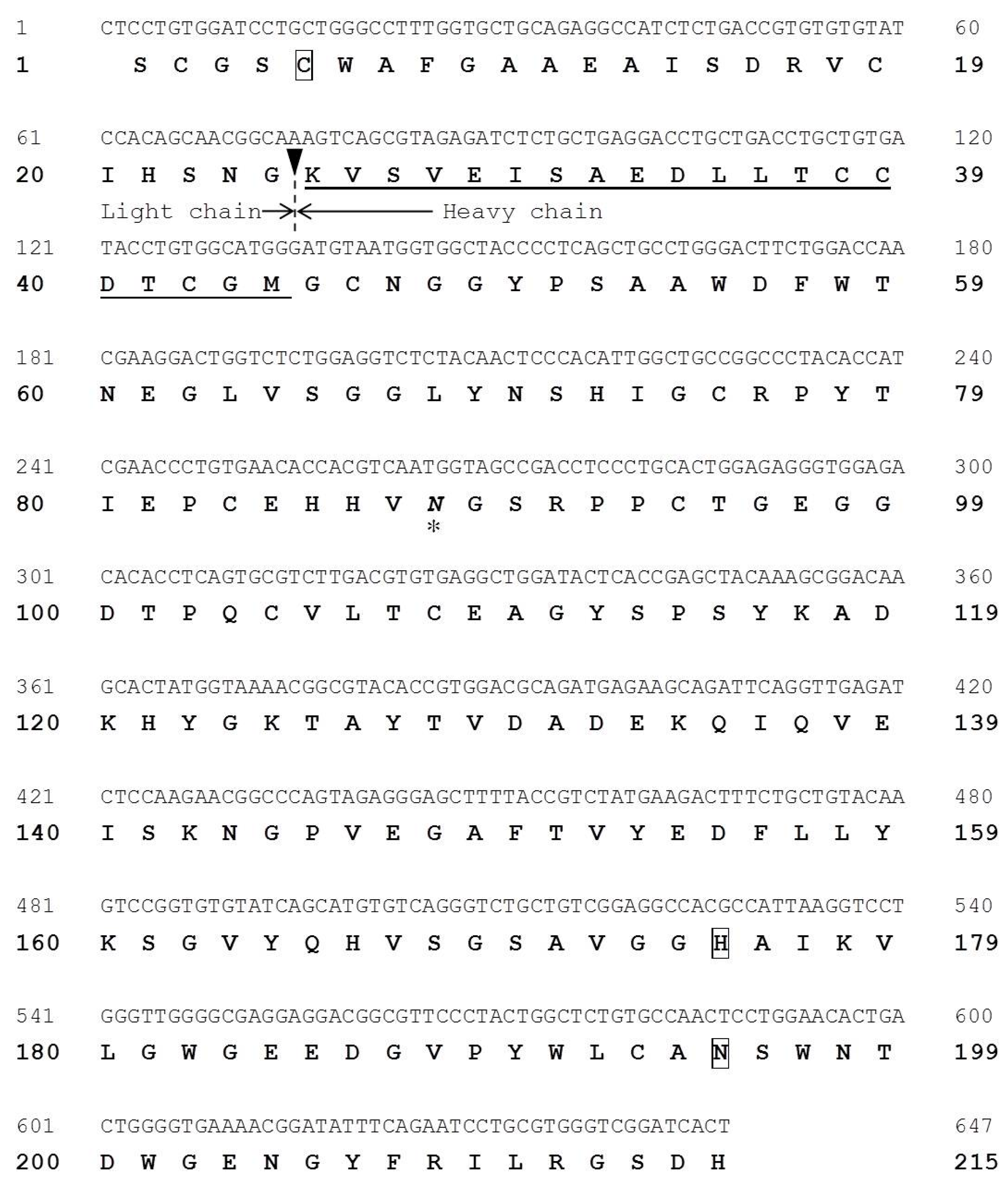

2.6. Effects of Cathepsin B Specific Inhibitors on Modori Gel
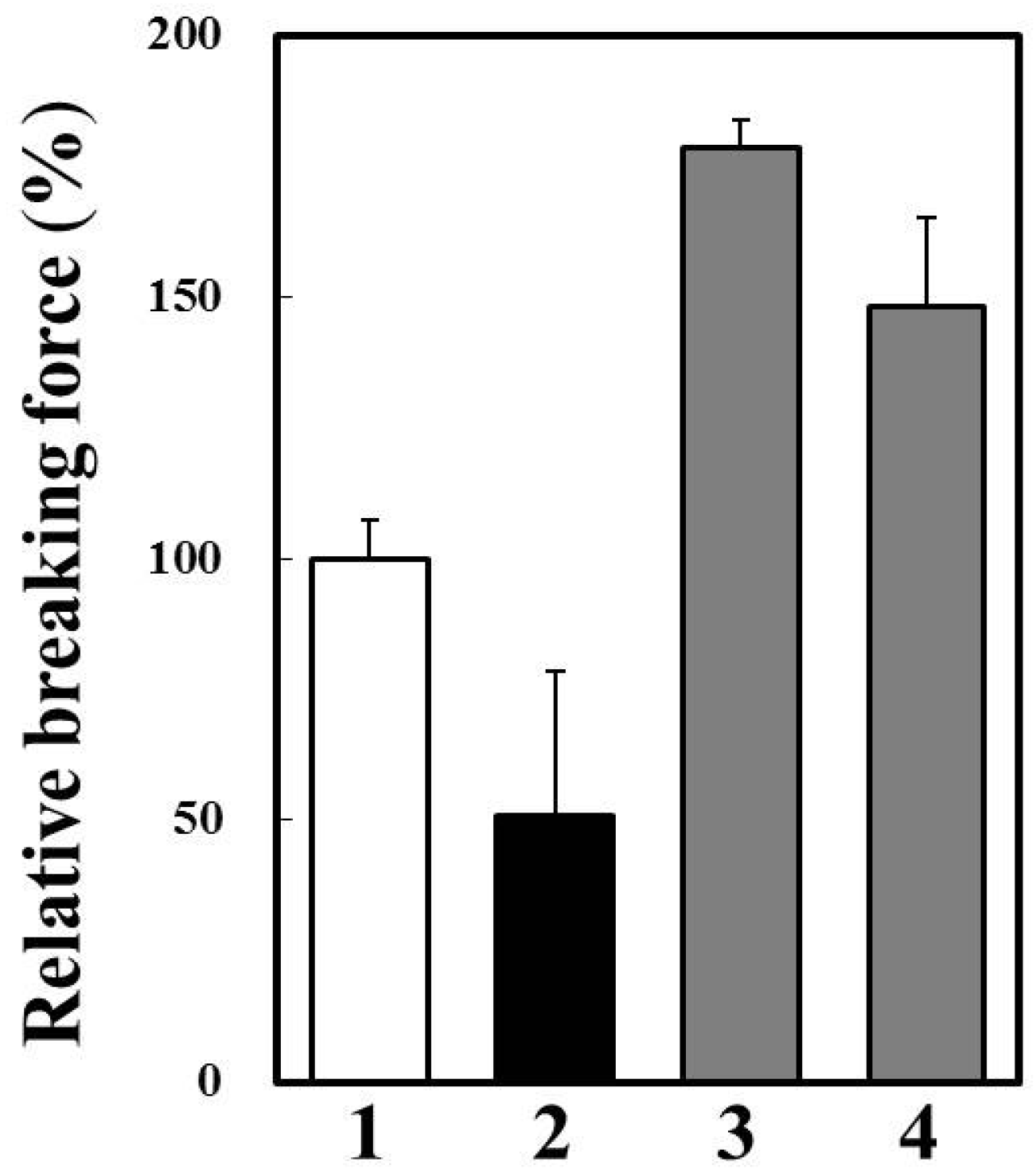
3. Experimental Section
3.1. Fish
3.2. Chemicals
3.3. Assay of Protease Activity
3.4. Purification of Cathepsin B
3.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
3.6. Measurement of Protein Concentration
3.7. Determination of N-terminal Amino Acid Sequence
3.8. Total RNA Isolation and cDNA Cloning by RT-PCR
3.9. DNA Sequencing and Analysis
3.10. Preparation and Evaluation of Thermal Gel
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mort, J.S.; Buttle, D.J. Cathepsin B. Int. J. Biochem. Cell Biol. 1997, 29, 715–720. [Google Scholar] [CrossRef]
- Sikorski, Z.; Kolakowski, E. Endogenous enzyme activity and seafood quality: Influence of chilling, freezing, and other environmental factors. In Seafood Enzymes: Utilization and Influence on Postharvest Seafood Quality; Haard, N.F., Simpson, B.K., Eds.; Marcel Dekker: NY, USA, 2000; pp. 451–487. [Google Scholar]
- Caballero, M.J.; Betancor, M.; Escrig, J.C.; Montero, D.; Espinosa de los Monteros, A.; Castro, P.; Ginés, R.; Izquierdo, M. Post mortem changes produced in the muscle of sea bream (Sparus aurata) during ice storage. Aquaculture 2009, 291, 210–216. [Google Scholar] [CrossRef]
- Yoshida, A.; Kurihara, M.; Ogata, H.; Cao, M.J.; Osatomi, K.; Hara, K. Proteolytic degradation of myofibrillar components by endogenous proteases in red sea bream muscle. Jpn. J. Food Chem. 2014, 21, 107–114. [Google Scholar]
- Yamashita, M.; Konagaya, S. Hydrolytic action of salmon cathepsin B and L to muscle structural proteins in respect of muscle softening. Nippon Suisan Gakkaishi 1991, 57, 1917–1922. [Google Scholar] [CrossRef]
- Aoki, T.; Ueno, R. Involvement of cathepsins B and L in the post-mortem autolysis of mackerel muscle. Food Res. Int. 1997, 30, 585–591. [Google Scholar] [CrossRef]
- Hara, K.; Suzumatsu, A.; Ishihara, T. Purification and characterization of cathepsin B from carp ordinary muscle. Nippon Suisan Gakkaishi 1988, 54, 1243–1252. [Google Scholar] [CrossRef]
- Liu, H.; Yin, L.; Zhang, N.; Li, S.; Ma, C. Isolation of cathepsin B from the muscle of silver carp (Hypophthalmichthys molitrix) and comparison of cathepsins B and L actions on surimi gel softening. Food Chem. 2008, 110, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Sherekar, S.V.; Gore, M.S.; Ninjoor, V. Purification and characterization of cathepsin B from the skeletal muscle of fresh water fish, Tilapia mossambica. J. Food Sci. 1988, 53, 1018–1023. [Google Scholar] [CrossRef]
- Jiang, S.T.; Lee, J.J.; Chen, H.C. Purification and characterization of cathepsin B from ordinary muscle of mackerel (Scomber australasicus). J. Agric. Food Chem. 1994, 42, 1073–1079. [Google Scholar] [CrossRef]
- Yamashita, M.; Konagaya, S. Purification and characterization of cathepsin B from the white muscle of chum salmon, oncorhynchus keta. Comp. Biochem. Physiol. 1990, 96B, 733–737. [Google Scholar]
- An, H.; Peters, M.Y.; Seymour, T.A. Roles of endogenous enzymes in surimi gelation. Trends Food Sci. Technol. 1996, 7, 321–327. [Google Scholar] [CrossRef]
- An, H.; Weerasingh, V.; Seymour, T.A.; Morrissey, M.T. Cathepsin degradation of Pacific whiting surimi proteins. J. Food Sci. 1994, 59, 1013–1017. [Google Scholar] [CrossRef]
- Jiang, M.T.; Lee, B.L.; Tsao, C.Y.; Lee, J.J. Mackerel Cathepsins B and L effects on thermal degradation of surimi. J. Food Sci. 1997, 62, 310–315. [Google Scholar] [CrossRef]
- Osatomi, K.; Sasai, H.; Cao, M.J.; Hara, K.; Ishihara, T. Purification and characterization of myofibril-bound serine proteinase from carp Cyprinus carpio ordinary muscle. Comp. Biochem. Physiol. 1997, 116B, 183–190. [Google Scholar] [CrossRef]
- Ohkubo, M.; Miyagawa, K.; Osatomi, K.; Hara, K.; Nozaki, Y.; Ishihara, T. Purification and characterization of myofibril-bound serine protease from lizard fish (Saurida undosquamis) muscle. Comp. Biochem. Physiol. 2004, 137B, 139–150. [Google Scholar] [CrossRef]
- Ohkubo, M.; Osatomi, K.; Hara, K.; Ishihara, T.; Aranishi, F. A novel type of myofibril-bound serine protease from white croaker (Argyrosomus argentatus). Comp. Biochem. Physiol. 2005, 141B, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.J.; Weng, L.; Liu, G.M.; Hara, K.; Su, W.J. Partial purification and characterization of tropomyosin-bound serine proteinase from the skeletal muscle of yellow croaker (Pseudosciaena crocea). J. Food Biochem. 2007, 31, 343–355. [Google Scholar] [CrossRef]
- Cao, M.J.; Wu, L.L.; Hara, K.; Weng, L.; Su, W.J. Purification and characterization of a myofibril-bound serine proteinase from the skeletal muscle of silver carp. J. Food Biochem. 2005, 29, 533–546. [Google Scholar] [CrossRef]
- Cao, M.J.; Jiang, X.J.; Zhong, H.C.; Zhang, Z.J.; Su, W.J. Degradation of myofibrillar proteins by a myofibril-bound serine proteinase in the skeletal muscle of crucian carp (Carasius auratus). Food Chem. 2006, 94, 7–13. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kato, K. Intracellular transport and processing of lysosomal cathepsin B. Biochem. Biophys. Res. Commun. 1987, 148, 254–259. [Google Scholar] [CrossRef]
- Hara, K.; Kominami, E.; Katunuma, N. Effect of proteinase inhibitors on intracellular processing of cathepsin B, H and L in rat macrophages. FEBS Lett. 1988, 231, 229–231. [Google Scholar] [CrossRef]
- Zhong, C.; Cai, Q.F.; Liu, G.M.; Sun, L.C.; Hara, K.; Su, W.J.; Cao, M.J. Purification and characterization of cathepsin L from the skeletal muscle of blue scad (Decapterus maruadsi) and comparison of its role with myofibril-bound serine proteinase in the degradation of myofibrillar proteins. Food Chem. 2012, 133, 1560–1568. [Google Scholar] [CrossRef]
- Aranishi, F.; Hara, K.; Osatomi, K.; Ishihara, T. Purification and characterization of cathepsin B from hepatopancreas of carp cyprinus carpio. Comp. Biochem. Physiol. 1997, 117B, 579–587. [Google Scholar] [CrossRef]
- Abe, K.; Emori, Y.; Kondo, H.; Suzuki, K.; Arai, S. Molecular cloning of a cysteine proteinase inhibitor of rice (Oryzacystatin). J. Biol. Chem. 1987, 262, 16793–16797. [Google Scholar] [PubMed]
- Tanimoto, S.; Tomioka, M. Effects of rice-related products on the textural properties and color of fish meat gels derived from walleye pollock (Theragra chalcagramma). Food Sci. Technol. Res. 2013, 19, 463–470. [Google Scholar] [CrossRef]
- Tan, Y.; Osatomi, K.; Nozaki, Y.; Ishihara, T.; Hara, K. Occurrence of two distinct molecular species of cathepsin B in carp Cyprinus carpio. Fish. Sci. 2006, 72, 185–194. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Far, A.L.; Randall, R. Protein measurement with the Folin phenol regent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, A.; Ohta, M.; Kuwahara, K.; Cao, M.-J.; Hara, K.; Osatomi, K. Purification and Characterization of Cathepsin B from the Muscle of Horse Mackerel Trachurus japonicus. Mar. Drugs 2015, 13, 6550-6565. https://doi.org/10.3390/md13116550
Yoshida A, Ohta M, Kuwahara K, Cao M-J, Hara K, Osatomi K. Purification and Characterization of Cathepsin B from the Muscle of Horse Mackerel Trachurus japonicus. Marine Drugs. 2015; 13(11):6550-6565. https://doi.org/10.3390/md13116550
Chicago/Turabian StyleYoshida, Asami, Megumi Ohta, Koichi Kuwahara, Min-Jie Cao, Kenji Hara, and Kiyoshi Osatomi. 2015. "Purification and Characterization of Cathepsin B from the Muscle of Horse Mackerel Trachurus japonicus" Marine Drugs 13, no. 11: 6550-6565. https://doi.org/10.3390/md13116550