Gene Sequence Based Clustering Assists in Dereplication of Pseudoalteromonas luteoviolacea Strains with Identical Inhibitory Activity and Antibiotic Production
Abstract
:1. Introduction
2. Results and Discussion
2.1. Pseudoalteromonas luteoviolacea Production of Antibacterial Compounds and Antibacterial Activity
| Live cell inhibition | Supernatant inhibition | Antibacterial compounds produced | Inhibition profile | Genotype * | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | V. ang. | S. aureus | V. ang. | S. aureus | Violacein | PBP | Indolmycin | Chemotype | |||||
| 2ta16 | x | x | - | x | x | x | - | 1 | A | I | |||
| NCIMB 1944 | x | x | - | x | x | x | - | 1 | A | I | |||
| DSM6061 T | x | x | - | x | x | x | - | 1 | A | I | |||
| CPMOR-2 | x | x | - | x | x | x | - | 1 | A | I | |||
| S2607 | x | x | - | x | x | x | - | 1 | A | III | |||
| S4060-1 | x | x | - | x | x | x | - | 1 | A | III | |||
| S4047-1 | x | x | x | x | x | - | x | 2 | B | II | |||
| S4054 WT | x | x | x | x | x | - | x | 2 | B | II | |||
| CPMOR-1 | x | x | x | x | x | - | x | 2 | B | II | |||
| H33 | x | x | - | - | x | - | - | 3 | C | III | |||
| H33S | x | x | - | - | x | - | - | 3 | C | III | |||
| NCIMB 1942 | - | - | - | - | x | - | - | 3 | D | III | |||
| NCIMB 2035 | - | - | - | - | x | - | - | 3 | D | III | |||
2.2. Clustering of P. luteoviolacea Strains in Genotypes
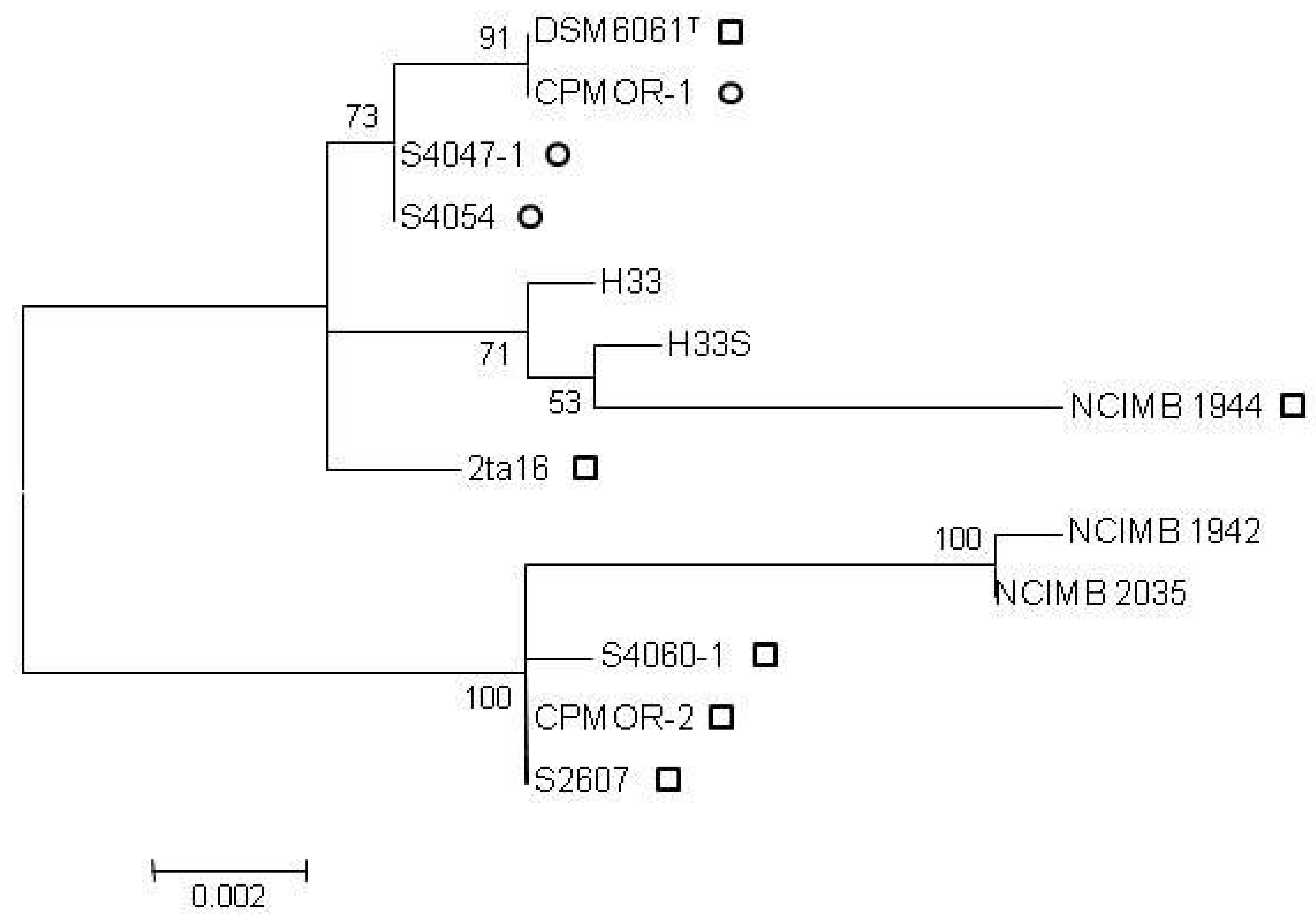

2.3. MIC of Known Antagonistic Compounds
3. Experimental Section
3.1. Bacterial Strains and Culture Conditions
| Strain name | Origin | Source |
|---|---|---|
| DSM 6061 T | Mediterranean, Nice | Surface water |
| S2607 | Pacific, Eastern Australia | Rock surface |
| S4060-1 | Pacific, Costa Rica | Seaweed |
| 2ta16 | Florida Keys, USA | M. annularis coral |
| CPMOR-2 | Mediterranian, Murcia | Surface water |
| NCIMB1944 | Mediterranean, Nice | Surface water |
| S4047-1 | Pacific, Costa Rica | Seaweed |
| S4054 | Pacific, Costa Rica | Seaweed |
| CPMOR-1 | Mediterranean, Murcia | Macroalgae |
| H33 | Sydney, Australia | Unknown |
| H33S | Sydney, Australia | Unknown |
| NCIMB1942 | Mediterranean, Nice | Surface water |
| NCIMB2035 | Mediterranean, Nice | Surface water |
3.2. Assays for Inhibition of Bacterial Growth
3.3. Analytical Detection of Antibacterial Compounds
3.4. PCR Amplification and Sequencing
3.5. Phylogenetic Analysis
3.6. Minimum Inhibitory Concentrations (MIC) of Violacein and Indolmycin
4. Conclusions
Acknowledgments
References
- Newman, D.J. Natural products as leads to potential drugs: An old process or the new hope for drug discovery? J. Med. Chem. 2008, 51, 2589–2599. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef]
- Molinari, G. Natural Products in Drug Discovery: Present Status and Perspectives. In Pharmaceutical Biotechnology; Guzmán, C.A., Feuerstein, G.Z., Eds.; Springer: New York, NY, USA, 2009; pp. 13–27. [Google Scholar]
- Baker, D.D.; Chu, M.; Oza, U. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef]
- Fenical, W. Chemical studies of marine bacteria—developing a new resource. Chem. Rev. 1993, 93, 1673–1683. [Google Scholar]
- Long, R.A.; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microb. 2001, 67, 4975–4983. [Google Scholar] [CrossRef]
- Gulder, T.A.M.; Moore, B.S. Chasing the treasures of the sea—bacterial marine natural products. Curr. Opin. Microbiol. 2009, 12, 252–260. [Google Scholar]
- Taori, K.; Paul, V.J.; Luesch, H. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am. Chem. Soc. 2008, 130, 1806–1807. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Harrison, S.T. Total Synthesis of Abyssomicin C and atrop-Abyssomicin C. Angew. Chem. Int. Ed. 2006, 45, 3256–3260. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Bach, G.; Heidorn, T.; Liang, L.; Schlingloff, A.; Simon, M. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microb. 2004, 70, 2560–2565. [Google Scholar]
- Hjelm, M.; Bergh, Ø.; Riaza, A.; Nielsen, J.; Melchiorsen, J.; Jensen, S.; Duncan, H.; Ahrens, P.; Birkbeck, H.; Gram, L. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 2004, 27, 360–371. [Google Scholar] [CrossRef]
- Bowman, J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef]
- Vynne, N.G.; Månsson, M.; Nielsen, K.F.; Gram, L. Bioactivity, chemical profiling and 16S rRNA based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar. Biotechnol. 2011, 13, 1062–1073. [Google Scholar] [CrossRef]
- Wietz, M.; Gram, L.; Jørgensen, B.; Schramm, A. Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 2010, 61, 179–189. [Google Scholar]
- Mikhailov, V.V.; Romanenko, L.A.; Ivanova, E.P. The genus Alteromonas and related Proteobacteria. In The Prokaryotes, 3rd; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2002; pp. 597–645. [Google Scholar]
- Corley, D.G.; Durley, R.C. Strategies for Database Dereplication of Natural Products. J. Nat. Prod. 1994, 57, 1484–1490. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Rosaiah, J.N. Various dereplication strategies using LC-MS for rapid natural product lead identification and drug discovery. Nat. Prod. Commun. 2007, 2, 193–202. [Google Scholar]
- Hook, D.J.; Pack, E.J.; Yacobucci, J.J.; Guss, J. Approaches to automating the dereplication of bioactive natural products—the key step in high throughput screening of bioactive materials from natural sources. J. Biomol. Screen. 1997, 2, 145–152. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Månsson, M.; Rank, C.; Frisvad, J.C.; Larsen, T.O. Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 2011, 74, 2338–2348. [Google Scholar] [CrossRef]
- Larsen, T.O.; Smedsgaard, J.; Nielsen, K.F.; Hansen, M.E.; Frisvad, J.C. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 2005, 22, 672–695. [Google Scholar] [CrossRef]
- Smedsgaard, J.; Nielsen, J. Metabolite profiling of fungi and yeast: from phenotype to metabolome by MS and informatics. J. Exp. Bot. 2005, 56, 273–286. [Google Scholar] [CrossRef]
- Knight, V.; Sanglier, J.J.; DiTullio, D.; Braccili, S.; Bonner, P.; Waters, J.; Hughes, D.; Zhang, L. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 2003, 62, 446–458. [Google Scholar] [CrossRef]
- Porsby, C.H.; Nielsen, K.F.; Gram, L. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microb. 2008, 74, 7356–7364. [Google Scholar] [CrossRef]
- Jensen, P.R.; Williams, P.G.; Oh, D.C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microb. 2007, 73, 1146–1152. [Google Scholar] [CrossRef]
- Goodfellow, M.; Fiedler, H.P. A guide to successful bioprospecting: Informed by actinobacterial systematics. Antonie Van Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef]
- Månsson, M.; Phipps, R.K.; Gram, L.; Munro, M.H.; Larsen, T.O.; Nielsen, K.F. Explorative Solid-Phase Extraction (E-SPE) for Accelerated Microbial Natural Product Discovery. J. Nat. Prod. 2010, 73, 1126–1132. [Google Scholar] [CrossRef]
- Lichstein, H.C.; Vandesand, V.F. Violacein, an antibiotic pigment produced by Chromobacterium violaceum. J. Infect. Dis. 1945, 76, 47–51. [Google Scholar] [CrossRef]
- Marsh, W.S.; Garretson, A.L.; Wesel, E.M. PA 155 A, B, and X. Antibiotics produced by a strain of Streptomyces albus. Antibiot. Chemother. 1960, 10, 316–320. [Google Scholar]
- Burkholder, P.R.; Pfister, R.M.; Leitz, F.H. Production of a pyrrole antibiotic by a marine bacterium. Appl. Environ. Microb. 1966, 14, 649–653. [Google Scholar]
- Gauthier, M.J.; Flatau, G.N. Antibacterial activity of marine violet-pigmented Alteromonas with special reference to the production of brominated compounds. Can. J. Microbiol. 1976, 22, 1612–1619. [Google Scholar] [CrossRef]
- Matz, C.; Webb, J.S.; Schupp, P.J.; Phang, S.Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE 2008, 3, e2744. [Google Scholar]
- Kanamaru, T.; Nakano, Y.; Toyoda, Y.; Miyagawa, K.I.; Tada, M.; Kaisho, T.; Nakao, M. In vitro and in vivo antibacterial activities of TAK-083, an agent for treatment of Helicobacter pylori infection. Antimicrob. Agents Chemother. 2001, 45, 2455–2459. [Google Scholar] [CrossRef]
- von Wittenau, M.S.; Els, H. The structure of indolmycin. J. Am. Chem. Soc. 1961, 83, 4678–4680. [Google Scholar] [CrossRef]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I. Anti-staphylococcal activity of indolmycin, a potential topical agent for control of staphylococcal infections. J. Antimicrob. Chemother. 2004, 54, 549–552. [Google Scholar] [CrossRef]
- Gomez, D.; Espinosa, E.; Bertazzo, M.; Lucas-Elio, P.; Solano, F.; Sanchez-Amat, A. The macromolecule with antimicrobial activity synthesized by Pseudoalteromonas luteoviolacea strains is an L-amino acid oxidase. Appl. Microbiol. Biotechnol. 2008, 79, 925–930. [Google Scholar] [CrossRef]
- Rossello-Mora, R.; Amann, R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001, 25, 39–67. [Google Scholar]
- Coenye, T.; Gevers, D.; Peer, Y.V.; Vandamme, P.; Swings, J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 2005, 29, 147–167. [Google Scholar]
- Hilario, E.; Buckley, T.R.; Young, J.M. Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek 2004, 86, 51–64. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kasai, H.; Arnold, D.L.; Jackson, R.W.; Vivian, A.; Harayama, S. Phylogeny of the genus Pseudomonas: Intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 2000, 146, 2385–2394. [Google Scholar]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef]
- Egan, S.; Wiener, P.; Kallifidas, D.; Wellington, E.M.H. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Van Leeuwenhoek 2001, 79, 127–133. [Google Scholar] [CrossRef]
- Ginolhac, A.; Jarrin, C.; Robe, P.; Perrière, G.; Vogel, T.M.; Simonet, P.; Nalin, R. Type I polyketide synthases may have evolved through horizontal gene transfer. J. Mol. Evol. 2005, 60, 716–725. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T.; Clardy, J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. USA 2008, 105, 4601–4608. [Google Scholar]
- Strohl, W.R. Antimicrobials. In Microbial Diversity and Bioprospecting; Bull, A.T., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 36–355. [Google Scholar]
- Jensen, P.R. Linking species concepts to natural product discovery in the post-genomic era. J. Ind. Microbiol. Biotechnol. 2010, 37, 219–224. [Google Scholar] [CrossRef]
- Östling, J.; Goodman, A.E.; Kjelleberg, S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: Isolation of starvation inducible lac operon fusions. FEMS Microbiol. Ecol. 1991, 86, 83–94. [Google Scholar]
- Skov, M.N.; Pedersen, K.; Larsen, J.L. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl. Environ. Microb. 1995, 61, 1540–1545. [Google Scholar]
- Yamamoto, S.; Harayama, S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microb. 1995, 61, 1104–1109. [Google Scholar]
- Pascual, J.; Macián, M.C.; Arahal, D.R.; Garay, E.; Pujalte, M.J. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Bacteriol. 2010, 60, 154–165. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Clinical and Laboratory Standards Institute, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard; CLSI document M7-A7, 7th ed; Clinical and Laboratory Standards Institute: Pennsylvania, PA, USA, 2006.
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vynne, N.G.; Mansson, M.; Gram, L. Gene Sequence Based Clustering Assists in Dereplication of Pseudoalteromonas luteoviolacea Strains with Identical Inhibitory Activity and Antibiotic Production. Mar. Drugs 2012, 10, 1729-1740. https://doi.org/10.3390/md10081729
Vynne NG, Mansson M, Gram L. Gene Sequence Based Clustering Assists in Dereplication of Pseudoalteromonas luteoviolacea Strains with Identical Inhibitory Activity and Antibiotic Production. Marine Drugs. 2012; 10(8):1729-1740. https://doi.org/10.3390/md10081729
Chicago/Turabian StyleVynne, Nikolaj G., Maria Mansson, and Lone Gram. 2012. "Gene Sequence Based Clustering Assists in Dereplication of Pseudoalteromonas luteoviolacea Strains with Identical Inhibitory Activity and Antibiotic Production" Marine Drugs 10, no. 8: 1729-1740. https://doi.org/10.3390/md10081729




