Nanomaterial-Assisted Signal Enhancement of Hybridization for DNA Biosensors: A Review
Abstract
:1. Introduction
2. DNA Hybridization and Signal-Detecting Methods
3. Hybridization Signal Enhanced by Nanomaterials
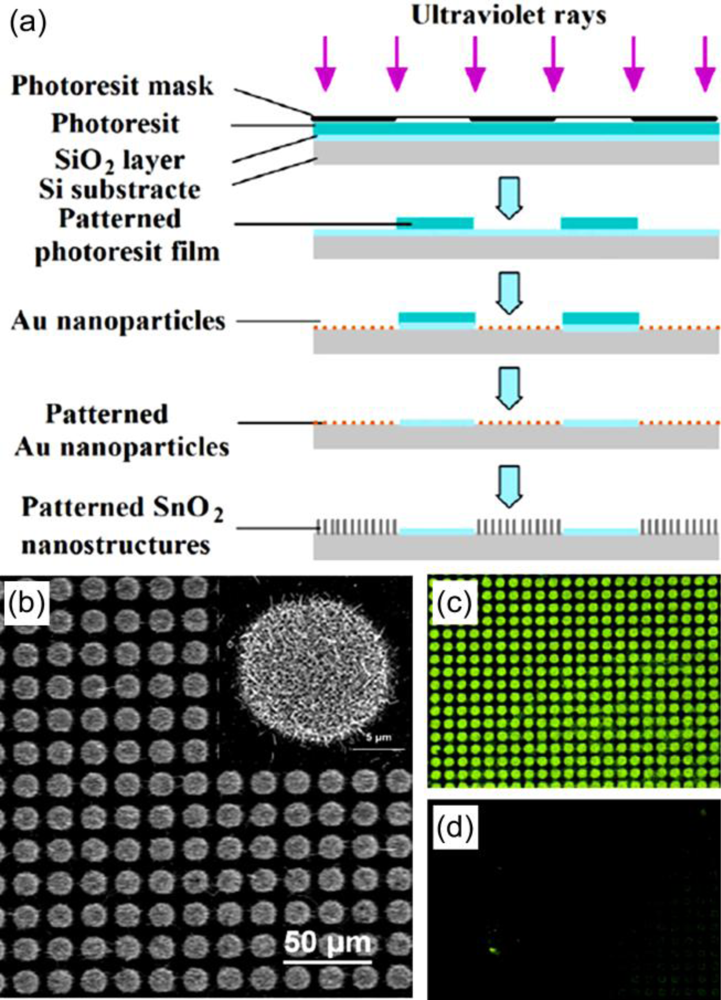
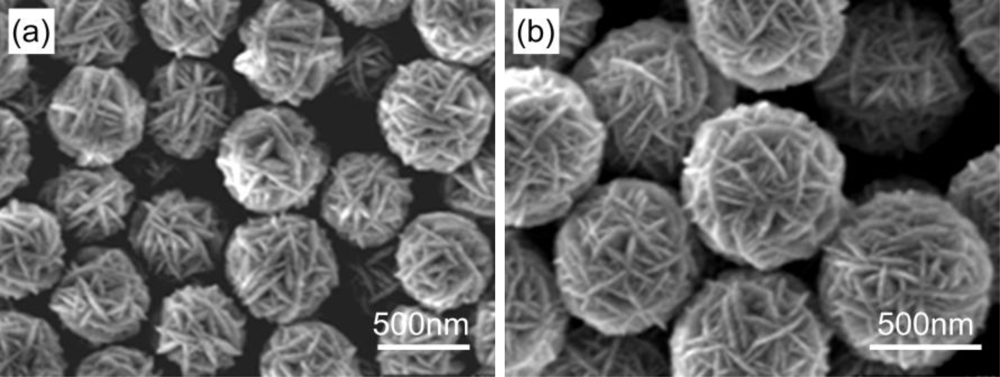
3.1. Nanosemiconductors
3.2. Quantum-dots (QDs)
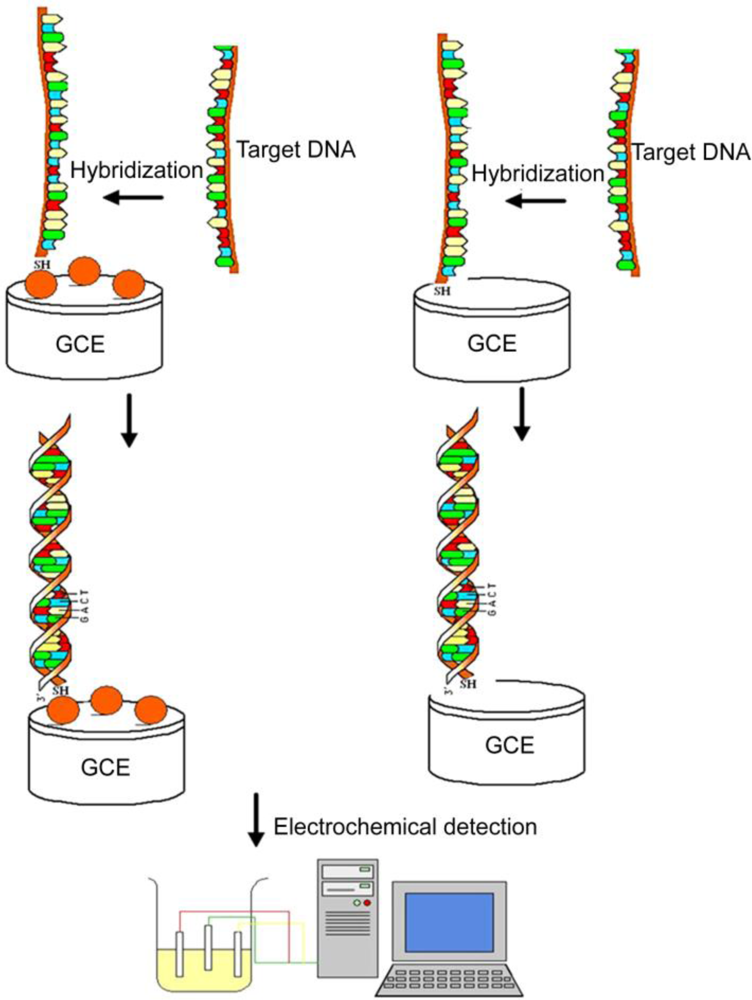
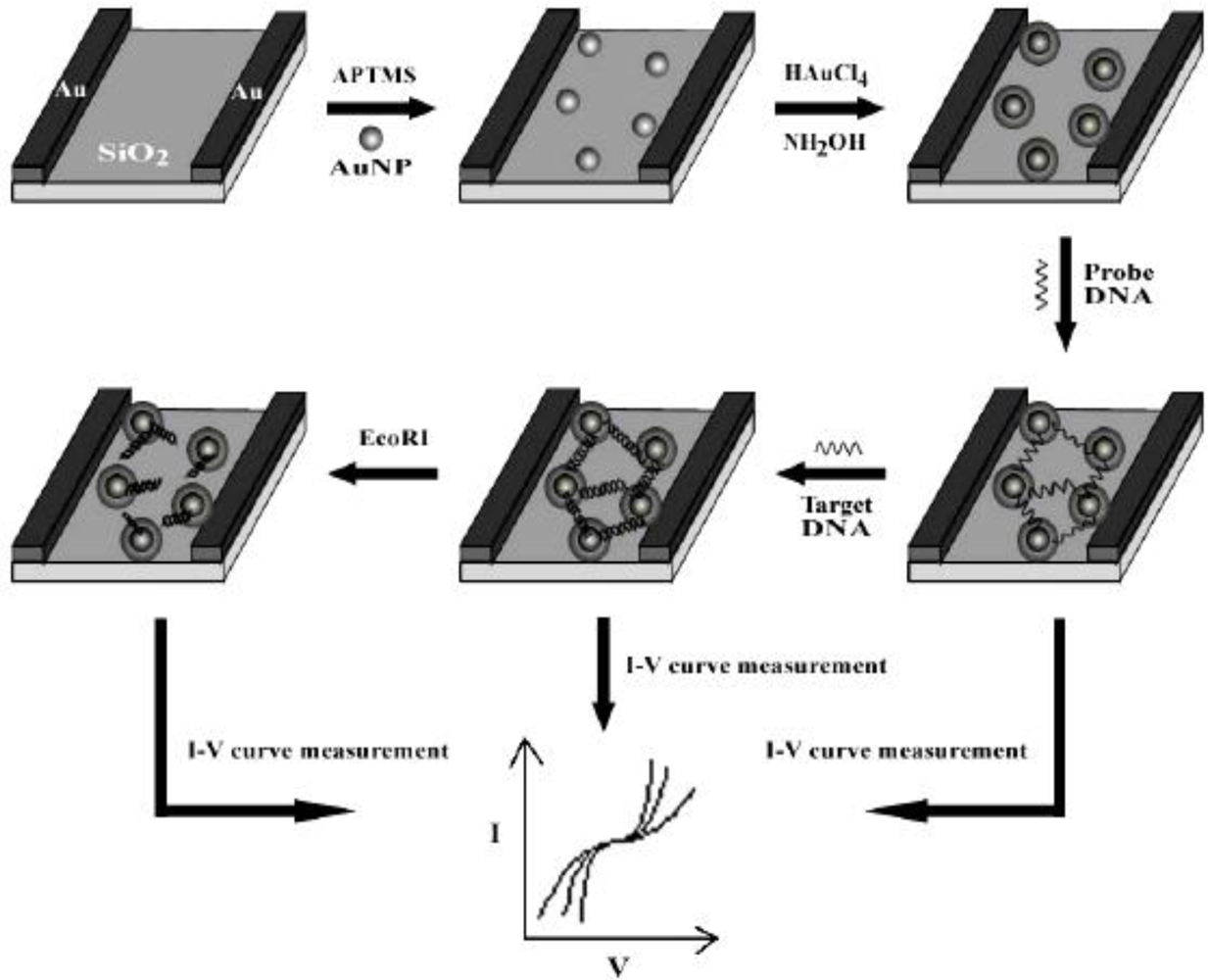
3.3. Nanoparticles (NPs)
3.4. Carbon nanotubes (CNTs) and their composites
4. Signal Enhancement of DNA Biosensor Based on Non-Nanomaterials for Comparison
5. Conclusions and Perspectives
Acknowledgments
References and Notes
- McCarty, M. Discovering genes are made of DNA. Nature 2003, 421, 406. [Google Scholar]
- Bostick, M.; Kim, J.K.; Esteve, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007, 317, 1760–1764. [Google Scholar]
- Kuwayama, H.; Yanagida, T.; Ueda, M. DNA oligonucleotide-assisted genetic manipulation increases transformation and homologous recombination efficiencies: Evidence from gene targeting of dictyostelium discoideum. J. Biotechnol 2008, 133, 418–423. [Google Scholar]
- Ainsworth, C. Disasters drive DNA forensics to reunite families. Nature 2006, 7094, 673. [Google Scholar]
- Guo, G.; Tong, T.Y. Genetic, demographic, and socioeconomic influences on timing of first sexual intercourse: Twin and DNA data of add health. Behav. Genet 2005, 35, 803. [Google Scholar]
- Alt, A.; Lammens, K.; Chiocchini, C.; Lammens, A.; Pieck, J.C.; Kuch, D.; Hopfner, K.P.; Carell, T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase. Science 2007, 318, 967–970. [Google Scholar]
- Borentain, P.; Gerolami, V.; Ananian, P.; Garcia, S.; Noundou, A.; Botta-Fridlund, D.; Le Treut, Y.P.; Berge-Lefranc, J.L.; Gerolami, R. DNA-repair and carcinogen-metabolising enzymes genetic polymorphisms as an independent risk factor for hepatocellular carcinoma in Caucasian liver-transplanted patients. Eur. J. Cancer 2007, 43, 2479–2486. [Google Scholar]
- Ferrando, A.; Lecis, R.; Domingo-Roura, X.; Ponsa, M. Genetic diversity and individual identification of reintroduced otters (Lutra lutra) in north-eastern Spain by DNA genotyping of spraints. Conserv. Genet 2008, 9, 129–139. [Google Scholar]
- Douglas, C.; Ehlting, J. Arabidopsis thaliana full genome longmer microarrays: a powerful gene discovery tool for agriculture and forestry. Transgenic Res 2005, 14, 551–561. [Google Scholar]
- Wu, J.; Prausnitz, J. Generalizations for the potential of mean force between two isolated colloidal particles from monte carlo simulations. J. Colloid Interface Sci 2002, 252, 326–330. [Google Scholar]
- Wang, S.; Cheng, Q. Microarray analysis in drug discovery and clinical applications. Methods Mol. Biol 2005, 316, 49–65. [Google Scholar]
- Kumar, N.; Dorfman, A.; Hahm, J. Ultrasensitive DNA sequence detection using nanoscale ZnO sensor arrays. Nanotechlology 2006, 17, 2875–2881. [Google Scholar]
- Kerman, K.; Kobayashi, M.; Tamiya, E. Recent trends in electrochemical DNA biosensor technology. Meas. Sci. Technol 2004, 15, R1–R11. [Google Scholar]
- Lagerqvist, J.; Zwolak, M.; Ventra, M. Fast DNA sequencing via transverse electronic transport. Nano Lett 2006, 6, 779–782. [Google Scholar]
- Lazerges, M.; Perrot, H.; Zeghib, N.; Antoine, E.; Compere, C. In situ QCM DNA-biosensor probe modification. Sens. Actuat. B 2006, 120, 329–337. [Google Scholar]
- Idegami, K.; Chikae, M.; Kerman, K.; Nagatani, N.; Yuhi, T.; Endo, T.; Tamiya, E. Gold nanoparticle-based redox signal enhancement for sensitive detection of human chorionic gonadotropin hormone. Electroanalysis 2008, 20, 14–21. [Google Scholar]
- He, Y.; Wang, H.F.; Yan, X.P. Self-assembly of Mn-doped ZnS quantum dots/octa(3-aminopropyl)octasilsequioxane octahydrochloride nanohybrids for optosensing DNA. Chem. Eur. J 2009, 15, 5436–5440. [Google Scholar]
- Malic, L.; Cui, B.; Veres, T.; Tabrizian, M. Enhanced surface plasmon resonance imaging detection of DNA hybridization on periodic gold nanoposts. Optics Lett 2007, 32, 3092–3094. [Google Scholar]
- Negulyaev, N.N.; Stepanyuk, V.S.; Niebergall, L.; Bruno, P.; Hergert, W.; Repp, J.; Rieder, K.H.; Meyer, G. Direct evidence for the effect of quantum confinement of surface-state electrons on atomic diffusion. Phys. Rev. Lett 2008, 101, 226601. [Google Scholar]
- Cheregi, F.S.; Vinattieri, A.; Feltin, E.; Simeonov, D.; Levrat, J.; Carlin, J.F.; Butte, R.; Grandjean, N.; Gurioli, M. Impact of quantum confinement and quantum confined Stark effect on biexciton binding energy in GaN/AlGaN quantum wells. Appl. Phys. Lett 2008, 93, 152105. [Google Scholar]
- Li, C.C.; Du, Z.F.; Yu, H.C.; Wang, T.H. Low-temperature sensing and high sensitivity of ZnO nanoneedles due to small size effect. Thin Solid Films 2009, 517, 5931–5934. [Google Scholar]
- Fagas, G.; Tkachov, G.; Pfund, A.; Richter, K. Geometrical enhancement of the proximity effect in quantum wires with extended superconducting tunnel contacts. Phys. Rev. B 2005, 71, 224510. [Google Scholar]
- Shevchenko, E.V.; Bodnarchitk, M.I.; Kovalenko, M.V.; Talapin, D.V.; Smith, R.K.; Aloni, S.; Heiss, W.; Alivisatos, A.P. Gold/iron oxide core/hollow-shell nanoparticles. Adv. Mater 2008, 20, 4323–4329. [Google Scholar]
- Wang, C.C.; Yu, C.Y.; Kei, C.C.; Lee, C.T.; Perng, T.P. The formation of TiO2 nanowires directly from nanoparticles. Nanotechnology 2009, 20, 285601. [Google Scholar]
- Yuen, M.Y.; Roy, V.A.L.; Lu, W.; Kui, S.C.F.; Tong, G.S.M.; So, M.H.; Chui, S.S.Y.; Muccini, M.; Ning, J.Q.; Xu, S.J.; Che, C.M. Semiconducting and electroluminescent nanowires self-assembled from organoplatinum(II) complexes. Angew. Chem. Int. Ed 2008, 47, 9895–9899. [Google Scholar]
- Tran, M.L.; Centeno, S.P.; Hutchison, J.A.; Engelkamp, H.; Liang, D.; Van Tendeloo, G.; Sels, B.F.; Hofkens, J.; Uji, I.H. Control of surface plasmon localization via self-assembly of silver nanoparticles along silver nanowires. J. Am. Chem. Soc 2008, 130, 17240–17241. [Google Scholar]
- Soci, C.; Bao, X.Y.; Aplin, D.P.R.; Wang, D.L. A systematic study on the growth of GaAs nanowires by metal-organic chemical vapor deposition. Nano Lett 2008, 8, 4275–4282. [Google Scholar]
- Maciel, I.O.; Delgado, J.C.; Silva, E.C.; Pimenta, M.A.; Sumpter, B.G.; Meunier, V.; Urias, F.L.; Sandoval, E.M.; Terrones, H.; Terrones, M.; Jorio, A. Synthesis, electronic structure, and raman scattering of phosphorus-doped single-wall carbon nanotubes. Nano Lett 2009, 9, 2267–2272. [Google Scholar]
- Li, W.Y.; Camargo, P.H.C.; Lu, X.M.; Xia, Y.N. Dimers of silver nanospheres: facile synthesis and their use as hot spots for surface-enhanced raman scattering. Nano Lett 2009, 9, 485–490. [Google Scholar]
- Lin, Y.W.; Huang, M.F.; Chang, H.T. Nanomaterials and chip-based nanostructures for capillary electrophoretic separations of DNA. Electrophoresis 2005, 26, 320–330. [Google Scholar]
- Healy, K. Nanopore-based single-molecule DNA analysis. Nanomedicine 2007, 2, 459–481. [Google Scholar]
- Wang, Z.D.; Lu, Y. Functional DNA directed assembly of nanomaterials for biosensing. J. Mater. Chem 2009, 19, 1788–1798. [Google Scholar]
- Wang, H.; Yang, R.H.; Yang, L.; Tan, W.H. Nucleic acid conjugated nanomaterials for enhanced molecular recognition. ACS NANOArticle ASAP. [CrossRef]
- Letuta, S.N.; Ketsle, G.A.; Levshin, L.V.; Nikiyan, A.N.; Davydova, O.K. A study of the interaction of rhodamine 6G with DNA by spectrophotometry and probe microscopy. Opt. Spectrosc 2002, 933, 844–847. [Google Scholar]
- Sadik, A.O.; Turner, J.L.H.; Kilgore, T.M.; Farkas, A.M.; Lotze, M.T. Comparative analysis of biolayer interferometry, small-volume spectrophotometry, and intercalating fluorometric assays for the detection of DNA in serum. J. Leukocyte Biol 2008, 84, A34. [Google Scholar]
- Matsumoto, Y.; Terui, N.; Tanaka, S. Electrochemical detection and control of interactions between DNA and electroactive intercalator using a DNA-Alginate complex film modified electrode. Environ. Sci. Technol 2006, 40, 4240–4244. [Google Scholar]
- Ferreyra, N.F.; Rivas, G.A. Self-assembled multilayers of polyethylenimine and DNA: spectrophotometric and electrochemical characterization and application for the determination of acridine orange interaction. Electroanalysis 2009, 21, 1665–1671. [Google Scholar]
- Nawaz, H.; Rauf, S.; Akhtar, K.; Khalid, A.M. Electrochemical DNA biosensor for the study of ciprofloxacin-DNA interaction. Anal. Biochem 2006, 354, 28–34. [Google Scholar]
- Li, F.; Chen, W.; Zhang, S.S. Development of DNA electrochemical biosensor based on covalent immobilization of probe DNA by direct coupling of sol-gel and self-assembly technologies. Biosens. Bioelectron 2008, 24, 781–786. [Google Scholar]
- Zhang, X.Z.; Jiao, K.; Liu, S.F.; Hu, Y.W. Readily reusable electrochemical DNA hybridization biosensor based on the interaction of DNA with single-walled carbon nanotubes. Anal. Chem 2009, 81, 6006–6012. [Google Scholar]
- Zhao, J.W.; Wu, L.Z.; Zhi, J.F. Fabrication of micropatterned ZnO/SiO2 core/shell nanorod arrays on a nanocrystalline diamond film and their application to DNA hybridization detection. J. Mater. Chem 2008, 18, 2459–2465. [Google Scholar]
- Lee, D.; Yoo, M.; Seo, H.; Tak, Y.; Kim, W.; Yong, K.; Rhee, S.; Jeon, S. Enhanced mass sensitivity of ZnO nanorod-grown quartz crystal microbalances. Sens. Actuat. B 2009, 135, 444–448. [Google Scholar]
- Gu, C.P.; Huang, J.R.; Ni, N.; Li, M.Q.; Liu, J.H. Detection of DNA hybridization based on SnO2 nanomaterial enhanced fluorescence. J. Phys. D: Appl. Phys 2008, 41, 175103. [Google Scholar]
- Mathur, S.; Erdemc, A.; Cavelius, C.; Bartha, S.; Altmayer, J. Amplified electrochemical DNA-sensing of nanostructured metal oxide films deposited on disposable graphite electrodes functionalized by chemical vapor deposition. Sens. Actuat. B 2009, 136, 432–437. [Google Scholar]
- Xu, Y.; Cai, H.; He, P.G.; Fang, Y.Z. Probing DNA hybridization by impedance measurement based on CdS-Oligonucleotide nanoconjugates. Electroanalysis 2004, 16, 150–155. [Google Scholar]
- Xia, Q.; Chen, X.; Liu, J.H. Cadmium sulfide-modified GCE for direct signal-amplified sensing of DNA hybridization. Biophys. Chem 2008, 136, 101–107. [Google Scholar]
- Hohng, S.; Ha, T. Single-molecule quantum-dot fluorescence resonance energy transfer. ChemPhysChem 2005, 6, 956–960. [Google Scholar]
- Willard, D.M.; Van Orden, A. Quantum dots: resonant energy-transfer sensor. Nat. Mater 2003, 2, 575–576. [Google Scholar]
- Wang, Y.; Zou, J.; Zhao, Z.M.; Hao, Z.; Wang, K.L. High quality InAs quantum dots grown on patterned Si with a GaAs buffer layer. Nanotechnology 2009, 20, 305301. [Google Scholar]
- Sun, J.; Wang, L.W.; Buhro, W.E. Synthesis of cadmium telluride quantum wires and the similarity of their effective band gaps to those of equidiameter cadmium telluride quantum dots. J. Am. Chem. Soc 2008, 130, 7997–8005. [Google Scholar]
- Ko, M.H.; Kim, S.; Kang, W.J.; Lee, J.H.; Kang, H.; Moon, S.H.; Hwang, D.W.; Ko, H.Y.; Lee, D.S. In vitro derby imaging of cancer biomarkers using quantum dots. Small 2009, 5, 1207–1212. [Google Scholar]
- Stopa, M.; Marcus, C.M. Magnetic field control of exchange and noise immunity in double quantum dots. Nano Lett 2009, 8, 1778–1782. [Google Scholar]
- Dayal, S.; Burda, C. Semiconductor quantum dots as two-photon sensitizers. J. Am. Chem. Soc 2008, 130, 2890–2891. [Google Scholar]
- Lu, S.; Madhukar, A. Nonradiative resonant excitation transfer from nanocrystal quantum dots to adjacent quantum channels. Nano Lett 2007, 7, 3443–3451. [Google Scholar]
- Wang, C.H.; Chen, T.T.; Tan, K.W.; Chen, Y.F.; Cheng, C.T.; Chou, P.T. Photoluminescence properties of CdTe/CdSe core-shell type-II quantumdots. J. Appl. Phys 2006, 99, 123521. [Google Scholar]
- Doose, S. Optical amplification from single excitons in collodial quantum dots. Small 2007, 3, 1856–1858. [Google Scholar]
- Duan, H.W.; Nie, S.M. Cell-penetrating quantum dots based on multivalent and endosome-disrupting surface coatings. J. Am. Chem. Soc 2007, 129, 3333–3338. [Google Scholar]
- Gopar, V.A.; Mendez-Bermudez, J.A.; Aly, A.H. Effects of Andreev reflection on the conductance of quantum chaotic dots. Phys. Rev. B 2009, 79, 245412. [Google Scholar]
- Dyadyusha, L.; Yin, H.; Jaiswal, S.; Brown, T.; Baumberg, J.J.; Booye, F.P.; Melvin, T. Quenching of CdSe quantum dot emission, a new approach for biosensing. Chem. Commun 2005, 3201–3203. [Google Scholar]
- Robelek, R.; Niu, L.F.; Schmid, E.L.; Knoll, W. Multiplexed hybridization detection of quantum dot-conjugated DNA sequences using surface plasmon enhanced fluorescence microscopy and spectrometry. Anal. Chem 2004, 76, 6160–6165. [Google Scholar]
- Ma, L.; Wu, S.M.; Huang, J.; Ding, Y.; Pang, D.W.; Li, L.J. Fluorescence in situ hybridization (FISH) on maize metaphase chromosomes with quantum dot-labeled DNA conjugates. Chromosoma 2008, 117, 181–187. [Google Scholar]
- Ho, Y.P.; Kung, M.C.; Yang, S.; Wang, T.H. Multiplexed hybridization detection with multicolor colocalization of quantum dot nanoprobes. Nano Lett 2005, 5, 1693–1697. [Google Scholar]
- Zhang, C.Y.; Yeh, H.C.; Kuroki, M.; Wang, T.H. Single-quantum-dot-based DNA nanosensor. Nature Mater 2005, 4, 826–831. [Google Scholar]
- Ko, S.H.; Grant, S.A. A novel FRET-based optical fiber biosensor for rapid detection of Salmonella typhimurium. Biosens. Bioelectron 2006, 21, 1283–1290. [Google Scholar]
- Willemse, M.; Janssen, E.; de Lange, F.; Wieringa, B.; Fransen, J. ATP and FRET-a cautionary note. Nature Biotechnol 2007, 25, 170–172. [Google Scholar]
- Liu, C.C.; Hang, H.Y. A progress in detection of interactions between macromolecules: Linked FRET using three color fluorophore. Progr. Biochem. Biophys 2006, 33, 292–296. [Google Scholar]
- Attar, H.A.; Monkman, A.P. Effect of surfactant on FRET and quenching in DNA sequence detection using conjugated polymers. Adv. Funct. Mater 2008, 18, 2498–2509. [Google Scholar]
- Padilla-Parra, S.; Auduge, N.; Coppey-Moisan, M.; Tramier, M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells. Biophys. J 2008, 95, 2976–2988. [Google Scholar]
- Koukiekolo, R.; Jakubek, Z.J.; Cheng, J.; Sagan, S.M.; Pezacki, J.P. Studies of a viral suppressor of RNA silencing p19-CFP fusion protein: a FRET-based probe for sensing double-stranded fluorophore tagged small RNAs. Biophys. Chem 2009, 143, 166–169. [Google Scholar]
- Miyake-Stoner, S.J.; Miller, A.M.; Hammill, J.T.; Peeler, J.C.; Hess, K.R.; Mehl, R.A.; Brewer, S.H. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry 2009, 48, 5953–5962. [Google Scholar]
- Peng, H.; Zhang, L.j.; Kjallman, T.H.M.; Soeller, C.; Sejdic, J.T. DNA hybridization detection with blue luminescent quantum dots and dye-labeled single-stranded DNA. J. Am. Chem. Soc 2007, 129, 3048–3049. [Google Scholar]
- Wu, C.S.; Cupps, J.M.; Fan, X.D. Compact quantum dot probes for rapid and sensitive DNA detection using highly efficient fluorescence resonant energy transfer. Nanotechnology 2009, 20, 305502. [Google Scholar]
- Kim, J.H.; Chaudhary, S.; Ozkan, M. Multicolour hybrid nanoprobes of molecular beacon conjugated quantum dots: FRET and gel electrophoresis assisted target DNA detection. Nanotechnology 2007, 18, 195105. [Google Scholar]
- Kim, Y.S.; Kim, B.C.; Lee, J.H.; Kim, J.B.; Gu, M.B. Specific detection of DNA using quantum dots and magnetic beads for large volume samples. Biotechnol. Bioprocess Eng 2006, 11, 449–454. [Google Scholar]
- Feng, C.L.; Zhong, X.H.; Steinhart, M.; Caminade, A.M.; Majoral, J.P.; Knoll, W. Graded-bandgap quantum-dot-modified nanotubes: a sensitive biosensor for enhanced detection of DNA hybridization. Adv. Mater 2007, 19, 1933–1936. [Google Scholar]
- Li, J.; Liu, Q.; Liu, Y.J.; Liu, S.C.; Yao, S.Z. DNA biosensor based on chitosan film doped with carbon nanotubes. Anal. Biochem 2005, 346, 107–114. [Google Scholar]
- Corgier, B.J.; Li, F.; Blum, L.J.; Marquette, C.A. On-chip chemiluminescent signal enhancement using nanostructured gold-modified carbon microarrays. Langmuir 2007, 23, 8619–8623. [Google Scholar]
- Shang, L.; Chen, H.J.; Deng, L.; Dong, S.J. Enhanced resonance light scattering based on biocatalytic growth of gold nanoparticles for biosensors design. Biosens. Bioelectron 2008, 23, 1180–1184. [Google Scholar]
- Wang, J.L.; Zhou, H.S. Aptamer-based Au nanoparticles-enhanced surface plasmon resonance detection of small molecules. Anal. Chem 2008, 80, 7174–7178. [Google Scholar]
- Chen, C.C.; Ko, F.H.; Chang, E.Y. Hybridization sensing by electrical enhancement with nanoparticles in nanogap. J. Vac. Sci. Technol. B 2008, 26, 2572–2577. [Google Scholar]
- Nie, L.B.; Yang, Y.; Li, S.; He, N.Y. Enhanced DNA detection based on the amplification of gold nanoparticles using quartz crystal microbalance. Nanotechnology 2007, 18, 305501. [Google Scholar]
- Lu, W.; Jin, Y.; Wang, G.; Chen, D.; Li, J.H. Enhanced photoelectrochemical method for linear DNA hybridization detection using Au-nanopaticle labeled DNA as probe onto titanium dioxide electrode. Biosens. Bioelectron 2008, 23, 1534–1539. [Google Scholar]
- Liu, S.F.; Li, Y.F.; Li, J.R.; Jiang, L. Enhancement of DNA immobilization and hybridization on gold electrode modified by nanogold aggregates. Biosens. Bioelectron 2005, 21, 789–795. [Google Scholar]
- Das, J.; Yang, H. Enhancement of electrocatalytic activity of DNA-conjugated gold nanoparticles and its application to DNA detection. J. Phys. Chem. C 2009, 113, 6093–6099. [Google Scholar]
- Liu, T.; Tang, J.; Jiang, L. The enhancement effect of gold nanoparticles as a surface modifier on DNA sensor sensitivity. Biochem. Biophys. Res. Commun 2004, 313, 3–7. [Google Scholar]
- Wang, C.J.; Huang, J.R.; Wang, J.; Gu, C.P.; Wang, J.H.; Zhang, B.C.; Liu, J.H. Fabrication of the nanogapped gold nanoparticles film for direct electrical detection of DNA and EcoRI endonuclease. Colloids Surf. B 2009, 69, 99–104. [Google Scholar]
- Tang, D.P.; Yuan, R.; Chai, Y. Magnetic core-shell Fe3O4@Ag nanoparticles coated carbon paste interface for studies of carcinoembryonic antigen in clinical immunoassay. J. Phys. Chem. B 2006, 110, 11640–11646. [Google Scholar]
- Miao, L.; Ina, Y.; Tanemura, S.; Jiang, T.; Taneura, M.; Kaneko, K.; Toh, S.; Mori, Y. Fabrication and photochromic study of titanate nanotube loaded with silver nanoparticles. Sur. Sci 2007, 601, 2792–2799. [Google Scholar]
- Zhang, J.; Malicka, J.; Gryczynski, I.; Lakowicz, J.R. Oligonucleotide-displaced organic monolayer-protected silver nanoparticles and enhanced luminescence of their salted aggregates. Anal. Biochem 2004, 330, 81–86. [Google Scholar]
- Zhang, J.; Malicka, J.; Gryczynski, I.; Lakowicz, J.R. Surface-enhanced fluorescence of fluorescein-labeled oligonucleotides capped on silver nanoparticles. J. Phys. Chem. B 2005, 109, 7643–7648. [Google Scholar]
- Sabanayagam, C.R.; Lakowicz, J.R. Increasing the sensitivity of DNA microarrays by metal-enhanced fluorescence using surface-bound silver nanoparticles. Nucleic Acids Res 2007, 35. [Google Scholar] [CrossRef]
- Chen, Z.P.; Peng, Z.F.; Luo, Y.; Qu, B.; Jiang, J.H.; Zhang, X.B.; Shen, G.L.; Yu, R.Q. Successively amplified electrochemical immunoassay based on biocatalytic deposition of silver nanoparticles and silver enhancement. Biosens. Bioelectron 2007, 23, 485–491. [Google Scholar]
- Zhang, D.; Chen, Y.; Chen, H.Y.; Xia, X.H. Silica-nanoparticle-based interface for the enhanced immobilization and sequence-specific detection of DNA. Anal. Bioanal. Chem 2004, 379, 1025–1030. [Google Scholar]
- Iijima, S. Helical microtubules of graphitic carbon. Nature 1991, 354, 56–58. [Google Scholar]
- Rivas, G.A.; Pedano, M.L. Adsorption and electrooxidation of nucleic acids at carbon nanotubes paste electrodes. Electrochem. Commun 2004, 6, 10–16. [Google Scholar]
- Erdem, A.; Papakonstantinou, P.; Murphy, H. Direct DNA hybridization at disposable graphite electrodes modified with carbon nanotubes. Anal. Chem 2006, 78, 6656–6659. [Google Scholar]
- Wang, J.; Liu, G.D.; Jan, M.R. Ultrasensitive electrical biosensing of proteins and DNA: carbon-nanotube derived amplification of the recognition and transduction events. J. Am. Chem. Soc 2004, 126, 3010–3011. [Google Scholar]
- Niu, S.Y.; Zhao, M.; Hu, L.Z.; Zhang, S.S. Carbon nanotube-enhanced DNA biosensor for DNA hybridization detection using rutin-Mn as electrochemical indicator. Sens. Actuat. B 2008, 135, 200–205. [Google Scholar]
- Zhang, W.; Yang, T.; Huang, D.; Jiao, K.; Li, G.C. Synergistic effects of nano-ZnO/multi-walled carbon nanotubes/chitosan nanocomposite membrane for the sensitive detection of sequence-specific of PAT gene and PCR amplification of NOS gene. J. Membr. Sci 2008, 325, 245–251. [Google Scholar]
- Gu, C.P.; Huang, J.R.; Wang, J.H.; Wang, C.J.; Li, M.Q.; Liu, J.H. Enhanced electrochemical detection of DNA hybridization based on Au/MWCNTs nanocomposites. Anal. Lett 2007, 40, 1–11. [Google Scholar]
- Pappaert, K.; Vanderhoeven, J.; Van Hummelen, P.; Duttab, B.; Clicqa, D.; Barona, G.V.; Desmet, G. Enhancement of DNA micro-array analysis using a shear-driven micro-channel flow system. J. Chromatogr. A 2003, 1014, 1–9. [Google Scholar]
- Bynum, M.A.; Gordon, G.B. Hybridization enhancement using microfluidic planetary centrifugal mixing. Anal. Chem 2004, 76, 7039–7044. [Google Scholar]
- Brousseau, L.C. Label-free “digital detection” of single-molecule DNA hybridization with a single electron transistor. J. Am. Chem. Soc 2006, 128, 11346–11347. [Google Scholar]
- Yang, D.K.; Huang, J.L.; Chen, C.C.; Su, H.J.; Wu, J.C. Enhancement of target-DNA hybridization efficiency by pre-hybridization on sequence-orientated micro-arrayed probes. J. Chin. Inst. Chem. Eng 2008, 39, 187–193. [Google Scholar]
- Kojima, N.; Sugino, M.; Mikami, A.; Nonaka, K.; Fujinawa, Y.; Muto, I.; Matsubara, K.; Ohtsuka, E.; Komats, Y. Enhanced reactivity of amino-modified oligonucleotides by insertion of aromatic residue. Bioorg. Med. Chem. Lett 2006, 16, 5118–5121. [Google Scholar]
- Ren, X.S.; Xu, Q.H. Label-free DNA sequence detection with enhanced sensitivity and selectivity using cationic conjugated polymers and PicoGreen. Langmuir 2009, 25, 43–47. [Google Scholar]
- Woodyear, T.L.; Campbell, C.N.; Heller, A. Direct enzyme-amplified electrical recognition of a 30-base model oligonucleotide. J. Am. Chem. Soc 1996, 118, 5504–5505. [Google Scholar]
- Domıinguez, E.; Rincon, O.A.A.; Narvaez, A. Electrochemical DNA sensors based on enzyme dendritic architectures: an approach for enhanced sensitivity. Anal. Chem 2004, 76, 3132–3138. [Google Scholar]
- Roth, L.; Zagon, J.; Ehlers, A.; Kroh, L.W.; Broll, H. A novel approach for the detection of DNA using immobilized peptide nucleic acid (PNA) probes and signal enhancement by real-time immuno-polymerase chain reaction (RT-iPCR). Anal. Bioanal. Chem 2009, 394, 529–537. [Google Scholar]
- Li, X.M.; Zhan, Z.M.; Zhang, S.S.; Chen, H.Y. Enzyme enhanced quantitative determination of multiple DNA targets based on capillary electrophoresis. J. Chromatogr. A 2009, 1216, 2567–2573. [Google Scholar]
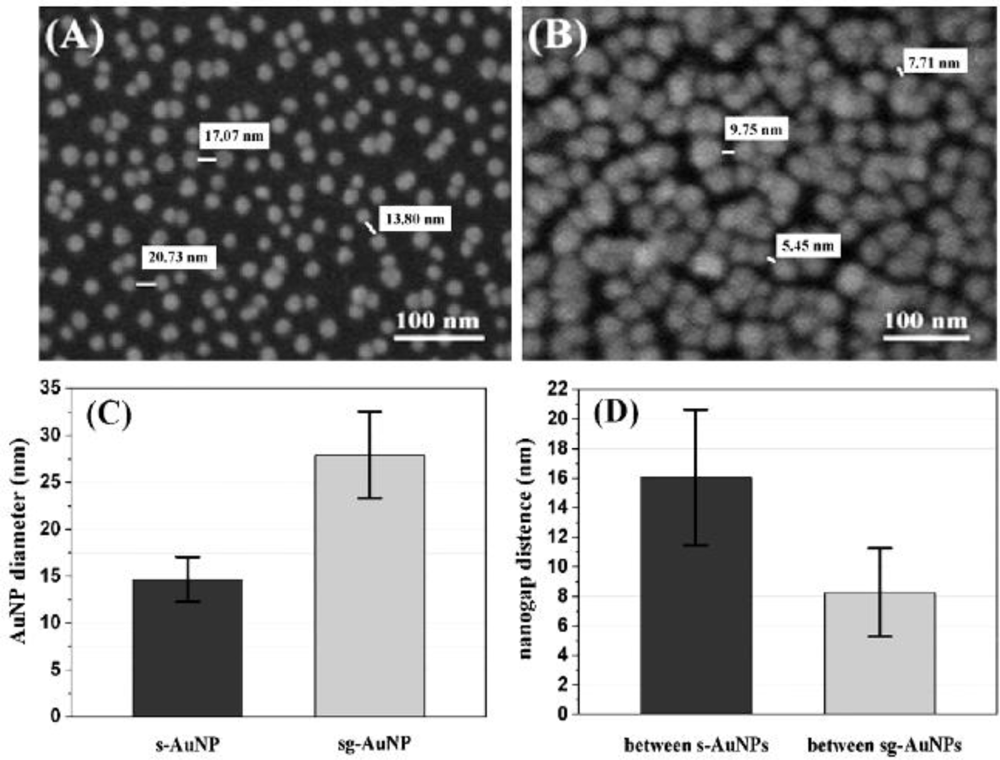
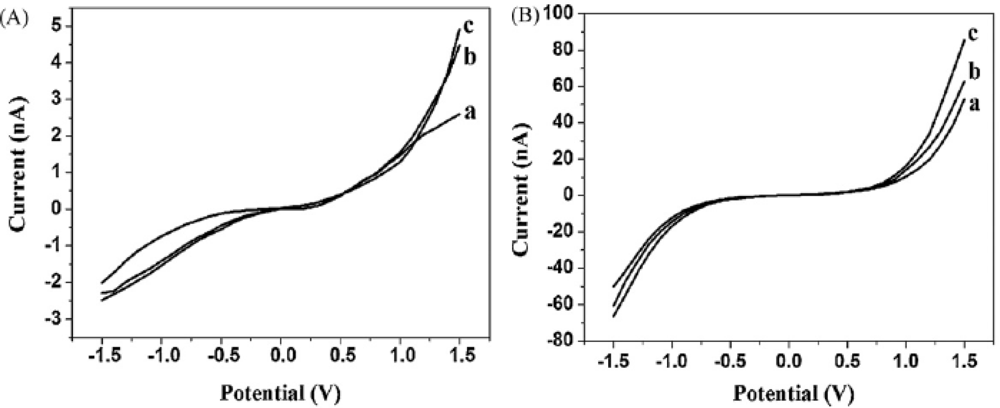

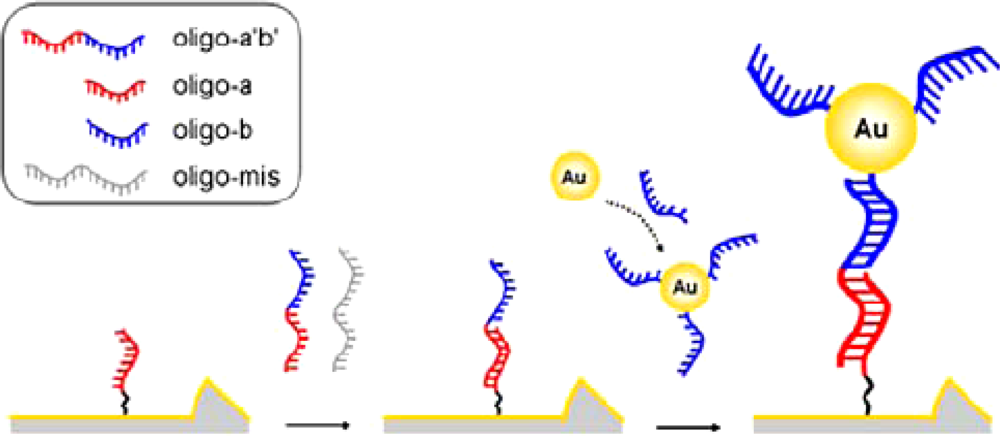
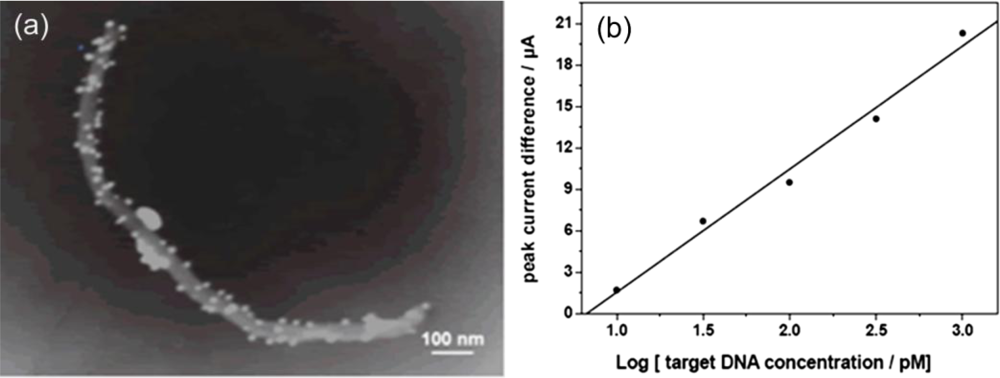
| Signal enhancers | Signal/method | Sensitivity enhancement | Detection limit (M) | Reference |
|---|---|---|---|---|
| SnO2 nanorods | fluorescence | 5-fold | 1.0 × 10−12 | 43 |
| nanoscale TiO2, SnO2, and Fe3O4 films | current | 2.1, 5.4, and 4.6 × 10−8, respectively | 44 | |
| walnut-like CdS NPs | current/cyclic and differential pulse voltammetry | ca. 3-fold | 6.0 × 10−14 | 46 |
| streptavidin-functionalized QDs | fluorescent/colorimetric measurement | < 1.0 × 10−9 | 62 | |
| single-QD | fluorescence/FRET | 100-fold | 4.8 × 10−15 | 63 |
| CdSe/ZnS core-shell QDs | fluorescence/FRET | < 1.0 × 10−9 | 72 | |
| CdSe-ZnS QDs and magnetic beads | fluorescence/FRET | >100-fold | 5.0 × 10−13 | 74 |
| ZnCdSe alloy QDs | fluorescence/FRET | ca. 15-fold | 75 | |
| AuNPs | frequency | 1.0 × 10−14 | 81 | |
| AuNPs | photocurrent | ca. 2-fold | 82 | |
| AuNPs | current/cyclic voltammetry | 1.0 × 10−11 | 83 | |
| AgNPs | fluorescence | >10-fold | 90 | |
| AgNPs | fluorescence | 28-fold | 91 | |
| AgNPs | current/anodic stripping voltammetry | 0.03 ng/mL (IgG) | 92 | |
| MWCNTs | current/cyclic voltammetry | 3.81 × 10−11 | 98 | |
| MWCNTs/ZnO/chitosan composites | current/differential pulse voltammetry | 2.8 × 10−12 | 99 | |
| Au/MWCNTs | current/differential pulse voltammetry | < 1.0 × 10−12 | 100 | |
| single electron transistor (non-nanomaterials) | current | 2.5 × 10−11 | 103 | |
| cationic conjugated polymers and PicoGreen (non-nanomaterials) | fluorescence | 1.0 × 10−10 | 106 | |
| enzyme | current | 2-fold | 1.0 × 10−9 | 108 |
| enzyme | current | 1.2 × 10−11 | 110 |
© 2009 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, J.; Liu, J.; Yang, L.; Chen, X.; Zhang, M.; Meng, F.; Luo, T.; Li, M. Nanomaterial-Assisted Signal Enhancement of Hybridization for DNA Biosensors: A Review. Sensors 2009, 9, 7343-7364. https://doi.org/10.3390/s90907343
Liu J, Liu J, Yang L, Chen X, Zhang M, Meng F, Luo T, Li M. Nanomaterial-Assisted Signal Enhancement of Hybridization for DNA Biosensors: A Review. Sensors. 2009; 9(9):7343-7364. https://doi.org/10.3390/s90907343
Chicago/Turabian StyleLiu, Jinhuai, Jinyun Liu, Liangbao Yang, Xing Chen, Meiyun Zhang, Fanli Meng, Tao Luo, and Minqiang Li. 2009. "Nanomaterial-Assisted Signal Enhancement of Hybridization for DNA Biosensors: A Review" Sensors 9, no. 9: 7343-7364. https://doi.org/10.3390/s90907343




