Gram-Negative Bacterial Sensors for Eukaryotic Signal Molecules
Abstract
:1. Introduction
2. Discussion
2.1. Gram-negative Bacteria and Neurotransmitters
2.2. Gram-negative Bacteria and Immune System Communication Molecules
2.3. Gram-negative Bacteria and Peptide Hormones
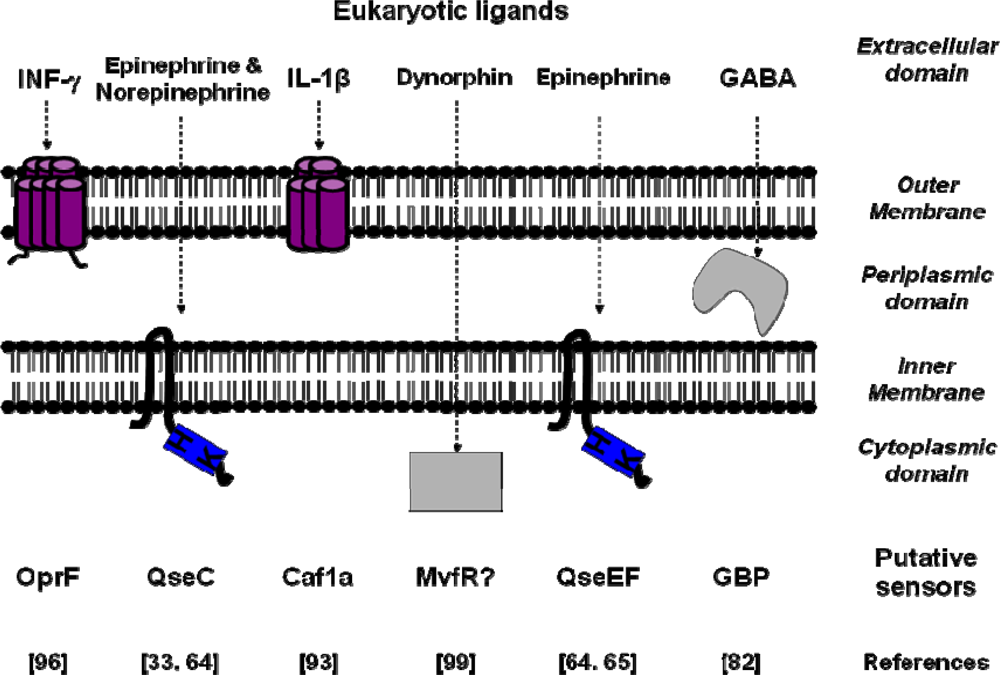
3. Conclusions and Perspectives
Acknowledgments
References and Notes
- Henderson, B.; Wilson, M. Homo bacteriens and a network of surprises. J. Med. Microbiol 1996, 45, 393–394. [Google Scholar]
- Honey, K. Good bugs, bad bugs: learning what we can from the microorganisms that colonize our bodies. J. Clin. Invest 2008, 118, 3817. [Google Scholar]
- Steinert, M.; Hentschel, U.; Hacker, J. Symbiosis and pathogenesis: evolution of the microbe-host interaction. Naturwissenschaften 2000, 87, 1–11. [Google Scholar]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar]
- Inglis, R.F.; Gardner, A.; Cornelis, P.; Buckling, A. Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 5703–5707. [Google Scholar]
- Rosenberger, C.M.; Finlay, B.B. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell. Biol 2003, 4, 385–396. [Google Scholar]
- Cossart, P.; Sansonetti, P.J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 2004, 304, 242–248. [Google Scholar]
- Wang, M.; Shakhatreh, M.A.; James, D.; Liang, S.; Nishiyama, S.; Yoshimura, F.; Demuth, D.R.; Hajishengallis, G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol 2007, 179, 2349–2358. [Google Scholar]
- Mitchell, J.A.; Paul-Clark, M.J.; Clarke, G.W.; McMaster, S.K.; Cartwright, N. Critical role of toll-like receptors and nucleotide oligomerisation domain in the regulation of health and disease. J. Endocrinol 2007, 193, 323–330. [Google Scholar]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol 2001, 1, 135–145. [Google Scholar]
- Lien, E.; Means, T.K.; Heine, H.; Yoshimura, A.; Kusumoto, S.; Fukase, K.; Fenton, M.J.; Oikawa, M.; Qureshi, N.; Monks, B.; et al. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Invest 2000, 105, 497–504. [Google Scholar]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem 2002, 71, 635–700. [Google Scholar]
- Schurr, J.R.; Young, E.; Byrne, P.; Steele, C.; Shellito, J.E.; Kolls, J.K. Central role of toll-like receptor 4 signaling and host defense in experimental pneumonia caused by Gram-negative bacteria. Infect. Immun 2005, 73, 532–545. [Google Scholar]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res 2003, 338, 2431–2447. [Google Scholar]
- Kawasaki, K.; Ernst, R.K.; Miller, S.I. Deacylation and palmitoylation of lipid A by Salmonellae outer membrane enzymes modulate host signaling through Toll-like receptor 4. J. Endotoxin Res 2004, 10, 439–444. [Google Scholar]
- Kawahara, K.; Tsukano, H.; Watanabe, H.; Lindner, B.; Matsuura, M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun 2002, 70, 4092–4098. [Google Scholar]
- Ernst, R.K.; Adams, K.N.; Moskowitz, S.M.; Kraig, G.M.; Kawasaki, K.; Stead, C.M.; Trent, M.S.; Miller, S.I. The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J. Bacteriol 2006, 188, 191–201. [Google Scholar]
- Padda, J.S.; Schryvers, A.B. N-linked oligosaccharides of human transferrin are not required for binding to bacterial transferrin receptors. Infect. Immun 1990, 58, 2972–2976. [Google Scholar]
- Ullberg, M.; Kronvall, G.; Karlsson, I.; Wiman, B. Receptors for human plasminogen on gram-negative bacteria. Infect. Immun 1990, 58, 21–25. [Google Scholar]
- Lopes, J.D.; dos Reis, M.; Brentani, R.R. Presence of laminin receptors in Staphylococcus aureus. Science 1985, 229, 275–277. [Google Scholar]
- Holderbaum, D.; Hall, G.S.; Ehrhart, L.A. Collagen binding to Staphylococcus aureus. Infect. Immun 1986, 54, 359–364. [Google Scholar]
- Visai, L.; Speziale, P.; Bozzini, S. Binding of collagens to an enterotoxigenic strain of Escherichia coli. Infect. Immun 1990, 58, 449–455. [Google Scholar]
- Rebiere-Huet, J.; Guerillon, J.; Pimenta, A.L.; Di Martino, P.; Orange, N.; Hulen, C. Porins of Pseudomonas fluorescens MFO as fibronectin-binding proteins. FEMS Microbiol. Lett 2002, 215, 121–126. [Google Scholar]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol 2000, 3, 165–170. [Google Scholar]
- Beier, D.; Gross, R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol 2006, 9, 143–152. [Google Scholar]
- Anantharaman, V.; Aravind, L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 2003, 4, 34. [Google Scholar]
- Nikolskaya, A.N.; Mulkidjanian, A.Y.; Beech, I.B.; Galperin, M.Y. MASE1 and MASE2: two novel integral membrane sensory domains. J. Mol. Microbiol. Biotechnol 2003, 5, 11–16. [Google Scholar]
- Zhulin, I.B.; Nikolskaya, A.N.; Galperin, M.Y. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol 2003, 185, 285–294. [Google Scholar]
- Tasneem, A.; Iyer, L.M.; Jakobsson, E.; Aravind, L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol 2005, 6, R4. [Google Scholar]
- Anantharaman, V.; Aravind, L. MEDS and PocR are novel domains with a predicted role in sensing simple hydrocarbon derivatives in prokaryotic signal transduction systems. Bioinformatics 2005, 21, 2805–2811. [Google Scholar]
- Galperin, M.Y.; Nikolskaya, A.N. Identification of sensory and signal-transducing domains in two-component signaling systems. Methods Enzymol 2007, 422, 47–74. [Google Scholar]
- Ravichandran, A.; Sugiyama, N.; Tomita, M.; Swarup, S.; Ishihama, Y. Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 2009, 9, 2764–2775. [Google Scholar]
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar]
- Lyte, M.; Ernst, S. Catecholamine induced growth of gram negative bacteria. Life Sci 1992, 50, 203–212. [Google Scholar]
- Lyte, M.; Ernst, S. Alpha and beta adrenergic receptor involvement in catecholamine-induced growth of gram-negative bacteria. Biochem. Biophys. Res. Commun 1993, 190, 447–452. [Google Scholar]
- Lyte, M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol 2004, 12, 14–20. [Google Scholar]
- Freestone, P.P.; Lyte, M. Microbial endocrinology: experimental design issues in the study of interkingdom signalling in infectious disease. Adv. Appl. Microbiol 2008, 64, 75–105. [Google Scholar]
- Freestone, P.P.; Sandrini, S.M.; Haigh, R.D.; Lyte, M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol 2008, 16, 55–64. [Google Scholar]
- Lyte, M.; Arulanandam, B.; Nguyen, K.; Frank, C.; Erickson, A.; Francis, D. Norepinephrine induced growth and expression of virulence associated factors in enterotoxigenic and enterohemorrhagic strains of Escherichia coli. Adv. Exp. Med. Biol 1997, 412, 331–339. [Google Scholar]
- Porat, R.; Clark, B.D.; Wolff, S.M.; Dinarello, C.A. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science 1991, 254, 430–432. [Google Scholar]
- Hogan, J.S.; Todhunter, D.A.; Smith, K.L.; Schoenberger, P.S.; Sordillo, L.M. Growth responses of coliform bacteria to recombinant bovine cytokines. J. Dairy Sci 1993, 76, 978–982. [Google Scholar]
- Woods, D.E.; Jones, A.L.; Hill, P.J. Interaction of insulin with Pseudomonas pseudomallei. Infect. Immun 1993, 61, 4045–4050. [Google Scholar]
- Yamashita, K.; Kaneko, H.; Yamamoto, S.; Konagaya, T.; Kusugami, K.; Mitsuma, T. Inhibitory effect of somatostatin on Helicobacter pylori proliferation in vitro. Gastroenterology 1998, 115, 1123–1130. [Google Scholar]
- Veron, W.; Lesouhaitier, O.; Pennanec, X.; Rehel, K.; Leroux, P.; Orange, N.; Feuilloley, M.G. Natriuretic peptides affect Pseudomonas aeruginosa and specifically modify lipopolysaccharide biosynthesis. FEBS J 2007, 274, 5852–5864. [Google Scholar]
- Renaud, M.; Miget, A. Rôle favorisant des perturbations locales causées par l'adrénaline sur le développement des infections microbiennes. C. R. Séances Soc. Biol. Fil 1930, 103, 1052–1054. [Google Scholar]
- Lyte, M. The role of microbial endocrinology in infectious disease. J. Endocrinol 1993, 137, 343–345. [Google Scholar]
- Freestone, P.P.; Haigh, R.D.; Williams, P.H.; Lyte, M. Stimulation of bacterial growth by heat-stable, norepinephrine-induced autoinducers. FEMS Microbiol. Lett 1999, 172, 53–60. [Google Scholar]
- Anderson, M.T.; Armstrong, S.K. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J. Bacteriol 2006, 188, 5731–5740. [Google Scholar]
- Lyte, M.; Arulanandam, B.P.; Frank, C.D. Production of Shiga-like toxins by Escherichia coli O157:H7 can be influenced by the neuroendocrine hormone norepinephrine. J. Lab. Clin. Med 1996, 128, 392–398. [Google Scholar]
- Lyte, M.; Erickson, A.K.; Arulanandam, B.P.; Frank, C.D.; Crawford, M.A.; Francis, D.H. Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun 1997, 232, 682–686. [Google Scholar]
- Lyte, M.; Freestone, P.P.; Neal, C.P.; Olson, B.A.; Haigh, R.D.; Bayston, R.; Williams, P.H. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet 2003, 361, 130–135. [Google Scholar]
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956. [Google Scholar]
- Rahman, H.; Reissbrodt, R.; Tschape, H. Effect of norepinephrine on growth of Salmonella and its enterotoxin production. Indian J. Exp. Biol 2000, 38, 285–286. [Google Scholar]
- Kinney, K.S.; Austin, C.E.; Morton, D.S.; Sonnenfeld, G. Catecholamine enhancement of Aeromonas hydrophila growth. Microb. Pathog 1999, 26, 85–91. [Google Scholar]
- O'Donnell, P.M.; Aviles, H.; Lyte, M.; Sonnenfeld, G. Enhancement of in vitro growth of pathogenic bacteria by norepinephrine: importance of inoculum density and role of transferrin. Appl. Environ. Microbiol 2006, 72, 5097–5099. [Google Scholar]
- Coulanges, V.; Andre, P.; Vidon, D.J. Effect of siderophores, catecholamines, and catechol compounds on Listeria spp. Growth in iron-complexed medium. Biochem. Biophys. Res. Commun 1998, 249, 526–530. [Google Scholar]
- Freestone, P.P.; Lyte, M.; Neal, C.P.; Maggs, A.F.; Haigh, R.D.; Williams, P.H. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol 2000, 182, 6091–6098. [Google Scholar]
- Freestone, P.P.; Haigh, R.D.; Lyte, M. Blockade of catecholamine-induced growth by adrenergic and dopaminergic receptor antagonists in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica. BMC Microbiol 2007, 7, 8. [Google Scholar]
- Freestone, P.P.; Haigh, R.D.; Lyte, M. Specificity of catecholamine-induced growth in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica. FEMS Microbiol. Lett 2007, 269, 221–228. [Google Scholar]
- Sperandio, V.; Li, C.C.; Kaper, J.B. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun 2002, 70, 3085–3093. [Google Scholar]
- Sperandio, V.; Torres, A.G.; Kaper, J.B. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol 2002, 43, 809–821. [Google Scholar]
- Hughes, D.T.; Sperandio, V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol 2008, 6, 111–120. [Google Scholar]
- Walters, M.; Sperandio, V. Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol 2006, 296, 125–131. [Google Scholar]
- Rasko, D.A.; Moreira, C.G.; Li de, R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC signaling and virulence for antibiotic development. Science 2008, 321, 1078–1080. [Google Scholar]
- Reading, N.C.; Rasko, D.A.; Torres, A.G.; Sperandio, V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 5889–5894. [Google Scholar]
- Bearson, B.L.; Bearson, S.M. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog 2008, 44, 271–278. [Google Scholar]
- Merighi, M.; Septer, A.N.; Carroll-Portillo, A.; Bhatiya, A.; Porwollik, S.; McClelland, M.; Gunn, J.S. Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol 2009, 9, 42. [Google Scholar]
- Belay, T.; Sonnenfeld, G. Differential effects of catecholamines on in vitro growth of pathogenic bacteria. Life Sci 2002, 71, 447–456. [Google Scholar]
- Alverdy, J.; Holbrook, C.; Rocha, F.; Seiden, L.; Wu, R.L.; Musch, M.; Chang, E.; Ohman, D.; Suh, S. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann. Surg 2000, 232, 480–489. [Google Scholar]
- Laughlin, R.S.; Musch, M.W.; Hollbrook, C.J.; Rocha, F.M.; Chang, E.B.; Alverdy, J.C. The key role of Pseudomonas aeruginosa PA-I lectin on experimental gut-derived sepsis. Ann. Surg 2000, 232, 133–142. [Google Scholar]
- Wagner, J.; Short, K.; Catto-Smith, A.G.; Cameron, D.J.; Bishop, R.F.; Kirkwood, C.D. Identification and characterisation of Pseudomonas 16S ribosomal DNA from ileal biopsies of children with Crohn's disease. PLoS ONE 2008, 3, e3578. [Google Scholar]
- Owens, D.F.; Kriegstein, A.R. Is there more to GABA than synaptic inhibition? Nat. Rev. Neurosci 2002, 3, 715–727. [Google Scholar]
- Dover, S.; Halpern, Y.S. Utilization of -aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J. Bacteriol 1972, 109, 835–843. [Google Scholar]
- Higuchi, T.; Hayashi, H.; Abe, K. Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J. Bacteriol 1997, 179, 3362–3364. [Google Scholar]
- Noe, F.F.; Nickerson, W.J. Metabolism of 2-pyrrolidone and gamma-aminobutyric acid by Pseudomonas aeruginosa. J. Bacteriol 1958, 75, 674–681. [Google Scholar]
- Scott, E.M.; Jakoby, W.B. Soluble gamma-aminobutyric-glutamic transaminase from Pseudomonas fluorescens. J. Biol. Chem 1959, 234, 932–936. [Google Scholar]
- Minuk, G.Y. Gamma-aminobutyric acid (GABA) production by eight common bacterial pathogens. Scand. J. Infect. Dis 1986, 18, 465–467. [Google Scholar]
- Mountfort, D.O.; Pybus, V. Regulatory Influences on the Production of Gamma-Aminobutyric Acid by a Marine Pseudomonad. Appl. Environ. Microbiol 1992, 58, 237–242. [Google Scholar]
- Morse, D.E.; Hooker, N.; Duncan, H.; Jensen, L. ggr-Aminobutyric Acid, a Neurotransmitter, Induces Planktonic Abalone Larvae to Settle and Begin Metamorphosis. Science 1979, 204, 407–410. [Google Scholar]
- Morse, D.E.; Duncan, H.; Hooker, N.; Baloun, A.; Young, G. GABA induces behavioral and developmental metamorphosis in planktonic molluscan larvae. Fed. Proc 1980, 39, 3237–3241. [Google Scholar]
- Guthrie, G.D.; Nicholson-Guthrie, C.S. gamma-Aminobutyric acid uptake by a bacterial system with neurotransmitter binding characteristics. Proc. Natl. Acad. Sci. USA 1989, 86, 7378–7381. [Google Scholar]
- Guthrie, G.D.; Nicholson-Guthrie, C.S.; Leary, H.L., Jr. A bacterial high-affinity GABA binding protein: isolation and characterization. Biochem. Biophys. Res. Commun 2000, 268, 65–68. [Google Scholar]
- Watanabe, M.; Maemura, K.; Kanbara, K.; Tamayama, T.; Hayasaki, H. GABA and GABA receptors in the central nervous system and other organs. Int. Rev. Cytol 2002, 213, 1–47. [Google Scholar]
- Morera, S.; Gueguen-Chaignon, V.; Raffoux, A.; Faure, D. Cloning, purification, crystallization and preliminary X-ray analysis of a bacterial GABA receptor with a Venus flytrap fold. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 2008, 64, 1153–1155. [Google Scholar]
- Chevrot, R.; Rosen, R.; Haudecoeur, E.; Cirou, A.; Shelp, B.J.; Ron, E.; Faure, D. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2006, 103, 7460–7464. [Google Scholar]
- Chou, H.T.; Kwon, D.H.; Hegazy, M.; Lu, C.D. Transcriptome analysis of agmatine and putrescine catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol 2008, 190, 1966–1975. [Google Scholar]
- Chapalain, A.; Chevalier, S.; Lesouhaitier, O.; Merieau, A.; Orange, N.; Feuilloley, M.G. Involvement of gamma-aminobutyric acid (GABA) in the regulation of adhesion and virulence in Pseudomonas fluorescens MF37 and Pseudomonas aeruginosa PAO1. Proceedings of the Swiss Molecular Microbiology: Microbial evolution and adaptation Symposium, Villar-sur-Ollon, The Switzerland, June 11–13, 2007; p. 1.
- Lambert, G.P. Stress-induced gastrointestinal barrier dysfunction and its inflammatory effects. J. Anim. Sci 2009, 87, E101–108. [Google Scholar]
- Denis, M.; Campbell, D.; Gregg, E.O. Interleukin-2 and granulocyte-macrophage colony-stimulating factor stimulate growth of a virulent strain of Escherichia coli. Infect. Immun 1991, 59, 1853–1856. [Google Scholar]
- Meduri, G.U.; Kanangat, S.; Stefan, J.; Tolley, E.; Schaberg, D. Cytokines IL-1beta, IL-6, and TNF-alpha enhance in vitro growth of bacteria. Am. J. Respir. Crit. Care Med 1999, 160, 961–967. [Google Scholar]
- Luo, G.; Niesel, D.W.; Shaban, R.A.; Grimm, E.A.; Klimpel, G.R. Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun 1993, 61, 830–835. [Google Scholar]
- Meduri, G.U. Clinical review: a paradigm shift: the bidirectional effect of inflammation on bacterial growth. Clinical implications for patients with acute respiratory distress syndrome. Crit. Care 2002, 6, 24–29. [Google Scholar]
- Zav'yalov, V.P.; Chernovskaya, T.V.; Navolotskaya, E.V.; Karlyshev, A.V.; MacIntyre, S.; Vasiliev, A.M.; Abramov, V.M. Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett 1995, 371, 65–68. [Google Scholar]
- Schall, T.J.; Lewis, M.; Koller, K.J.; Lee, A.; Rice, G.C.; Wong, G.H.; Gatanaga, T.; Granger, G.A.; Lentz, R.; Raab, H.; et al. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 1990, 61, 361–370. [Google Scholar]
- Van Delden, C.; Iglewski, B.H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis 1998, 4, 551–560. [Google Scholar]
- Wu, L.; Estrada, O.; Zaborina, O.; Bains, M.; Shen, L.; Kohler, J.E.; Patel, N.; Musch, M.W.; Chang, E.B.; Fu, Y.X.; et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science 2005, 309, 774–777. [Google Scholar]
- Risdahl, J.M.; Khanna, K.V.; Peterson, P.K.; Molitor, T.W. Opiates and infection. J. Neuroimmunol 1998, 83, 4–18. [Google Scholar]
- Peterson, P.K.; Molitor, T.W.; Chao, C.C. The opioid-cytokine connection. J. Neuroimmunol 1998, 83, 63–69. [Google Scholar]
- Zaborina, O.; Lepine, F.; Xiao, G.; Valuckaite, V.; Chen, Y.; Li, T.; Ciancio, M.; Zaborin, A.; Petroff, E.; Turner, J.R.; et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog 2007, 3, e35. [Google Scholar]
- Marinova, Z.; Vukojevic, V.; Surcheva, S.; Yakovleva, T.; Cebers, G.; Pasikova, N.; Usynin, I.; Hugonin, L.; Fang, W.; Hallberg, M.; et al. Translocation of dynorphin neuropeptides across the plasma membrane. A putative mechanism of signal transmission. J. Biol. Chem 2005, 280, 26360–26370. [Google Scholar]
- Xiao, G.; Deziel, E.; He, J.; Lepine, F.; Lesic, B.; Castonguay, M.H.; Milot, S.; Tampakaki, A.P.; Stachel, S.E.; Rahme, L.G. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol 2006, 62, 1689–1699. [Google Scholar]
- Pritchard, D.; Hooi, D.; Watson, E.; Chow, S.; Telford, G.; Bycroft, B.; Chhabra, S.R.; Harty, C.; Camara, M.; Diggle, S.P. Bacterial Quorum Sensing Signalling Molecules as Immune Modulators; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Hooi, D.S.; Bycroft, B.W.; Chhabra, S.R.; Williams, P.; Pritchard, D.I. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun 2004, 72, 6463–6470. [Google Scholar]
- Pritchard, D.I. Immune modulation by Pseudomonas aeruginosa quorum-sensing signal molecules. Int. J. Med. Microbiol 2006, 296, 111–116. [Google Scholar]
- Skindersoe, M.E.; Zeuthen, L.H.; Brix, S.; Fink, L.N.; Lazenby, J.; Whittall, C.; Williams, P.; Diggle, S.P.; Froekiaer, H.; Cooley, M.; et al. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol. Med. Microbiol 2009, 55, 335–345. [Google Scholar]
- Waterer, G.W.; Wunderink, R.G. Increasing threat of Gram-negative bacteria. Crit. Care Med 2001, 29, N75–81. [Google Scholar]
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol 2008, 6, 17–27. [Google Scholar]
- Hancock, R.E. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis 2001, 1, 156–164. [Google Scholar]
- Hancock, R.E.; Patrzykat, A. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disord 2002, 2, 79–83. [Google Scholar]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol 1999, 11, 23–27. [Google Scholar]
- Hancock, R.E.; Chapple, D.S. Peptide antibiotics. Antimicrob. Agents Chemother 1999, 43, 1317–1323. [Google Scholar]
- Bader, M.W.; Sanowar, S.; Daley, M.E.; Schneider, A.R.; Cho, U.; Xu, W.; Klevit, R.E.; Le Moual, H.; Miller, S.I. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 2005, 122, 461–472. [Google Scholar]
- Kraus, D.; Peschel, A. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol 2006, 306, 231–250. [Google Scholar]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol 2006, 4, 529–536. [Google Scholar]
- Peschel, A. How do bacteria resist human antimicrobial peptides? Trends Microbiol 2002, 10, 179–186. [Google Scholar]
- Moss, J.E.; Fisher, P.E.; Vick, B.; Groisman, E.A.; Zychlinsky, A. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol 2000, 2, 443–452. [Google Scholar]
- Llama-Palacios, A.; Lopez-Solanilla, E.; Poza-Carrion, C.; Garcia-Olmedo, F.; Rodriguez-Palenzuela, P. The Erwinia chrysanthemi phoP-phoQ operon plays an important role in growth at low pH, virulence and bacterial survival in plant tissue. Mol. Microbiol 2003, 49, 347–357. [Google Scholar]
- Macfarlane, E.L.; Kwasnicka, A.; Hancock, R.E. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 2000, 146, 2543–2554. [Google Scholar]
- Gooderham, W.J.; Gellatly, S.L.; Sanschagrin, F.; McPhee, J.B.; Bains, M.; Cosseau, C.; Levesque, R.C.; Hancock, R.E. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 2009, 155, 699–711. [Google Scholar]
- Cutuli, M.; Cristiani, S.; Lipton, J.M.; Catania, A. Antimicrobial effects of alpha-MSH peptides. J. Leukoc. Biol 2000, 67, 233–239. [Google Scholar]
- Metz-Boutigue, M.H.; Goumon, Y.; Lugardon, K.; Strub, J.M.; Aunis, D. Antibacterial peptides are present in chromaffin cell secretory granules. Cell. Mol. Neurobiol 1998, 18, 249–266. [Google Scholar]
- Metz-Boutigue, M.H.; Kieffer, A.E.; Goumon, Y.; Aunis, D. Innate immunity: involvement of new neuropeptides. Trends Microbiol 2003, 11, 585–592. [Google Scholar]
- Brogden, K.A.; Guthmiller, J.M.; Salzet, M.; Zasloff, M. The nervous system and innate immunity: the neuropeptide connection. Nat. Immunol 2005, 6, 558–564. [Google Scholar]
- Hansen, C.J.; Burnell, K.K.; Brogden, K.A. Antimicrobial activity of Substance P and Neuropeptide Y against laboratory strains of bacteria and oral microorganisms. J. Neuroimmunol 2006, 177, 215–218. [Google Scholar]
- El Karim, I.A.; Linden, G.J.; Orr, D.F.; Lundy, F.T. Antimicrobial activity of neuropeptides against a range of micro-organisms from skin, oral, respiratory and gastrointestinal tract sites. J. Neuroimmunol 2008, 200, 11–16. [Google Scholar]
- Kanai, K.; Kondo, E.; Kurata, T. Affinity and response of Burkholderia pseudomallei and Burkholderia cepacia to insulin. Southeast Asian J. Trop. Med. Public Health 1996, 27, 584–591. [Google Scholar]
- Simpson, A.J.; Wuthiekanun, V. Interaction of insulin with Burkholderia pseudomallei may be caused by a preservative. J. Clin. Pathol 2000, 53, 159–160. [Google Scholar]
- Currie, B. Pseudomonas pseudomallei-insulin interaction. Infect. Immun 1995, 63, 3745. [Google Scholar]
- Chowers, M.Y.; Keller, N.; Bar-Meir, S.; Chowers, Y. A defined human gastrin sequence stimulates the growth of Helicobacter pylori. FEMS Microbiol. Lett 2002, 217, 231–236. [Google Scholar]
- Chowers, M.Y.; Keller, N.; Tal, R.; Barshack, I.; Lang, R.; Bar-Meir, S.; Chowers, Y. Human gastrin: a Helicobacter pylori--specific growth factor. Gastroenterology 1999, 117, 1113–1118. [Google Scholar]
- Lamberts, S.W.; Krenning, E.P.; Reubi, J.C. The role of somatostatin and its analogs in the diagnosis and treatment of tumors. Endocr. Rev 1991, 12, 450–482. [Google Scholar]
- Hofland, L.J.; Lamberts, S.W. The pathophysiological consequences of somatostatin receptor internalization and resistance. Endocr. Rev 2003, 24, 28–47. [Google Scholar]
- Bulet, P.; Stocklin, R.; Menin, L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev 2004, 198, 169–184. [Google Scholar]
- Krause, A.; Liepke, C.; Meyer, M.; Adermann, K.; Forssmann, W.G.; Maronde, E. Human natriuretic peptides exhibit antimicrobial activity. Eur. J. Med. Res 2001, 6, 215–218. [Google Scholar]
- Kourie, J.I. Synthetic mammalian C-type natriuretic peptide forms large cation channels. FEBS Lett 1999, 445, 57–62. [Google Scholar]
- Kalra, P.R.; Clague, J.R.; Bolger, A.P.; Anker, S.D.; Poole-Wilson, P.A.; Struthers, A.D.; Coats, A.J. Myocardial production of C-type natriuretic peptide in chronic heart failure. Circulation 2003, 107, 571–573. [Google Scholar]
- Rudiger, A.; Gasser, S.; Fischler, M.; Hornemann, T.; von Eckardstein, A.; Maggiorini, M. Comparable increase of B-type natriuretic peptide and amino-terminal pro-B-type natriuretic peptide levels in patients with severe sepsis, septic shock, and acute heart failure. Crit. Care Med 2006, 34, 2140–2144. [Google Scholar]
- Post, F.; Weilemann, L.S.; Messow, C.M.; Sinning, C.; Munzel, T. B-type natriuretic peptide as a marker for sepsis-induced myocardial depression in intensive care patients. Crit. Care Med 2008, 36, 3030–3037. [Google Scholar]
- Vila, G.; Resl, M.; Stelzeneder, D.; Struck, J.; Maier, C.; Riedl, M.; Hulsmann, M.; Pacher, R.; Luger, A.; Clodi, M. Plasma NT-proBNP increases in response to LPS administration in healthy men. J. Appl. Physiol 2008, 105, 1741–1745. [Google Scholar]
- Veron, W.; Orange, N.; Feuilloley, M.G.; Lesouhaitier, O. Natriuretic peptides modify Pseudomonas fluorescens cytotoxicity by regulating cyclic nucleotides and modifying LPS structure. BMC Microbiol 2008, 8, 114. [Google Scholar]
- Wolfgang, M.C.; Lee, V.T.; Gilmore, M.E.; Lory, S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 2003, 4, 253–263. [Google Scholar]
- Lory, S.; Wolfgang, M.; Lee, V.; Smith, R. The multi-talented bacterial adenylate cyclases. Int. J. Med. Microbiol 2004, 293, 479–482. [Google Scholar]
- Smith, R.S.; Wolfgang, M.C.; Lory, S. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun 2004, 72, 1677–1684. [Google Scholar]
- Lu, J.; Bao, Q.; Wu, J.; Wang, H.; Li, D.; Xi, Y.; Wang, S.; Yu, S.; Qu, J. CSCDB: the cAMP and cGMP signaling components database. Genomics 2008, 92, 60–64. [Google Scholar]
- Galperin, M.Y. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol 2004, 6, 552–567. [Google Scholar]
- Papadopoulos, V.; Baraldi, M.; Guilarte, T.R.; Knudsen, T.B.; Lacapere, J.J.; Lindemann, P.; Norenberg, M.D.; Nutt, D.; Weizman, A.; Zhang, M.R.; et al. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci 2006, 27, 402–409. [Google Scholar]
- Yeliseev, A.A.; Kaplan, S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem 1995, 270, 21167–21175. [Google Scholar]
- Davey, M.E.; de Bruijn, F.J. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl. Environ. Microbiol 2000, 66, 5353–5359. [Google Scholar]
- Chapalain, A.; Chevalier, S.; Orange, N.; Murillo, L.; Papadopoulos, V.; Feuilloley, M.G. Bacterial ortholog of mammalian translocator protein (TSPO) with virulence regulating activity. PLoS ONE 2009, 4, e6096. [Google Scholar]
- Yeliseev, A.A.; Krueger, K.E.; Kaplan, S. A mammalian mitochondrial drug receptor functions as a bacterial “oxygen” sensor. Proc. Natl. Acad. Sci. USA 1997, 94, 5101–5106. [Google Scholar]
- Patel, N.J.; Zaborina, O.; Wu, L.; Wang, Y.; Wolfgeher, D.J.; Valuckaite, V.; Ciancio, M.J.; Kohler, J.E.; Shevchenko, O.; Colgan, S.P.; et al. Recognition of intestinal epithelial HIF-1alpha activation by Pseudomonas aeruginosa. Am. J. Physiol. Gastrointest. Liver Physiol 2007, 292, G134–142. [Google Scholar]
- Kohler, J.E.; Zaborina, O.; Wu, L.; Wang, Y.; Bethel, C.; Chen, Y.; Shapiro, J.; Turner, J.R.; Alverdy, J.C. Components of intestinal epithelial hypoxia activate the virulence circuitry of Pseudomonas. Am. J. Physiol. Gastrointest. Liver Physiol 2005, 288, G1048–1054. [Google Scholar]
- Cogan, T.A.; Thomas, A.O.; Rees, L.E.; Taylor, A.H.; Jepson, M.A.; Williams, P.H.; Ketley, J.; Humphrey, T.J. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 2007, 56, 1060–1065. [Google Scholar]
- Lacoste, A.; Jalabert, F.; Malham, S.K.; Cueff, A.; Poulet, S.A. Stress and stress-induced neuroendocrine changes increase the susceptibility of juvenile oysters (Crassostrea gigas) to Vibrio splendidus. Appl. Environ. Microbiol 2001, 67, 2304–2309. [Google Scholar]
- Nakano, M.; Takahashi, A.; Sakai, Y.; Nakaya, Y. Modulation of pathogenicity with norepinephrine related to the type III secretion system of Vibrio parahaemolyticus. J. Infect. Dis 2007, 195, 1248–1249. [Google Scholar]
- Nakano, M.; Takahashi, A.; Sakai, Y.; Kawano, M.; Harada, N.; Mawatari, K.; Nakaya, Y. Catecholamine-induced stimulation of growth in Vibrio species. Lett. Appl. Microbiol 2007, 44, 649–653. [Google Scholar]
- Kowalska, K.; Carr, D.B.; Lipkowski, A.W. Direct antimicrobial properties of substance P. Life Sci 2002, 71, 747–750. [Google Scholar]
| Signal molecules | Bacterial species | Observed effect | References |
|---|---|---|---|
| Norepinephrine | Acinetobacter lwoffii | ↑Growth | [47] |
| Bordetella bronchiseptica | ↑Growth | [48] | |
| Campylobacter jejuni | ↑Growth, virulence | [154] | |
| Citrobacter freundii | ↑Growth | [47] | |
| Hafnia alvei | ↑Growth | [47] | |
| Morganella morganii | ↑Growth | [47] | |
| Proteus mirabilis | ↑Growth | [47] | |
| Shigella sonnei | ↑Growth | [55] | |
| Vibrio splendidus | ↑Growth | [155] | |
| Vibrio parahaemolyticus | ↑Growth, virulence | [156, 157] | |
| Xanthomonas maltophilia | ↑Growth | [47] | |
| Catecholamines | Escherichia coli | ↑Growth | [34, 58] |
| Aeromonas hydrophila | ↑Growth | [54] | |
| Pseudomonas aeruginosa | ↑Growth, virulence | [47, 55, 68, 69] | |
| Salmonella enterica | ↑Growth, adhesion | [59] | |
| Yersinia enterocolitica | ↑Growth | [34, 59] | |
| GABA | Agrobacterium tumefaciens | ↑Virulence | [85] |
| Pseudomonas aeruginosa | ↑Virulence | [87] | |
| Signal molecules | Bacterial species | Observed effect | References |
|---|---|---|---|
| IL-1β | Escherichia coli | ↑Growth | [40] |
| Acinetobacter spp | ↑Growth | [90] | |
| TNF-α | Shigella flexneri | ↑Invasion | [91] |
| IL-2 | Escherichia coli | ↑Growth | [89] |
| GM-CSF | Escherichia coli | ↑Growth | [89] |
| IL-6 | Pseudomonas aeruginosa | ↑Growth | [90] |
| INF-γ | Klebsiella pneumoniae | ↑Growth | [41] |
| Escherichia coli | ↑Growth | [41] | |
| Pseudomonas aeruginosa | ↑QS/virulence | [96] | |
| Dynorphin | Pseudomonas aeruginosa | ↑QS/virulence | [99] |
| Signal molecules | Bacterial species | Observed effect | References |
|---|---|---|---|
| Substance P | Escherichia coli | Antibacterial | [125] |
| Pseudomonas aeruginosa | Antibacterial | [158] | |
| Insulin | Burkholderia pseudomalei | ↓Growth | [42] |
| CGRP | Escherichia coli | Antibacterial | [126] |
| Chromogranins | Escherichia coli | Antibacterial | [122] |
| Somatostatin | Helicobacter pylori | ↓Growth | [43] |
| hBNP | Pseudomonas aeruginosa | ↑Virulence | [44] |
| Pseudomonas fluorescens | ↑Virulence | [141] | |
| E. coli; P. aeruginosa | Antibacterial | [135] | |
| CNP | Pseudomonas fluorescens | ↑Virulence | [141] |
| Pseudomonas aeruginosa | ↑Virulence | [44] | |
| Gastrin | Helicobacter pylori | ↑Growth | [131] |
| ACTH | Vibrio splendidus | ↑Growth | [155] |
| Neuropeptide Y | Escherichia coli | Antibacterial | [125] |
| Signal molecules | Putative sensors | Binding Site affinity | Bacterial species | References |
|---|---|---|---|---|
| IL-1β | Caf1A | Kd=0.14 nM | Yersinia pestis | [93] |
| TNF-α | - | Kd=2.5 nM | Shigella flexneri | [91] |
| Epinephrine/Norepinephrine | QseC | Escherichia coli | [33] | |
| Epinephrine | QseE/QseF | Escherichia coli | [64,65] | |
| Somatostatin | SSRT2-like | Kd=0.3 nM | Helicobacter pylori | [43] |
| INF-γ | OprF | Pseudomonas aeruginosa | [96] | |
| Dynorphin | Mvfr | Pseudomonas aeruginosa | [99] | |
| GABA | GBP | Kd=65 nM | Pseudomonas fluorescens | [82] |
© 2009 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lesouhaitier, O.; Veron, W.; Chapalain, A.; Madi, A.; Blier, A.-S.; Dagorn, A.; Connil, N.; Chevalier, S.; Orange, N.; Feuilloley, M. Gram-Negative Bacterial Sensors for Eukaryotic Signal Molecules. Sensors 2009, 9, 6967-6990. https://doi.org/10.3390/s90906967
Lesouhaitier O, Veron W, Chapalain A, Madi A, Blier A-S, Dagorn A, Connil N, Chevalier S, Orange N, Feuilloley M. Gram-Negative Bacterial Sensors for Eukaryotic Signal Molecules. Sensors. 2009; 9(9):6967-6990. https://doi.org/10.3390/s90906967
Chicago/Turabian StyleLesouhaitier, Olivier, Wilfried Veron, Annelise Chapalain, Amar Madi, Anne-Sophie Blier, Audrey Dagorn, Nathalie Connil, Sylvie Chevalier, Nicole Orange, and Marc Feuilloley. 2009. "Gram-Negative Bacterial Sensors for Eukaryotic Signal Molecules" Sensors 9, no. 9: 6967-6990. https://doi.org/10.3390/s90906967




